8PUG
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7NQ9
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 2-benzyl-N-cyclopropyl-6-(1-methyl-1H-1,2,3-triazol-4-yl)isonicotinamide | | Descriptor: | 1,2-ETHANEDIOL, 6-benzyl-N4-((1r,3r)-3-hydroxycyclobutyl)-N2-methylpyridine-2,4-dicarboxamide, Bromodomain-containing protein 2 | | Authors: | Chung, C.W. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
4TKN
 
 | | Structure of the SNX17 FERM domain bound to the second NPxF motif of KRIT1 | | Descriptor: | Krev interaction trapped protein 1, Sorting nexin-17 | | Authors: | Stiegler, A.L, Zhang, R, Liu, W, Boggon, T.J. | | Deposit date: | 2014-05-27 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Determinants for Binding of Sorting Nexin 17 (SNX17) to the Cytoplasmic Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 289, 2014
|
|
6YWK
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
8FXN
 
 | | Bromodomain of CBP liganded with iCBP7 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, tert-butyl {(1R,4s)-4-[(5M)-2-[(2S)-1-(3-tert-butylphenyl)-6-oxopiperidin-2-yl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-benzimidazol-1-yl]cyclohexyl}carbamate | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks are sensitive to EP300/CBP bromodomain inhibition
To be published
|
|
1EGQ
 
 | | ENHANCEMENT OF ENZYME ACTIVITY THROUGH THREE-PHASE PARTITIONING: CRYSTAL STRUCTURE OF A MODIFIED SERINE PROTEINASE AT 1.5 A RESOLUTION | | Descriptor: | ACETIC ACID, CALCIUM ION, PROTEINASE K | | Authors: | Singh, R.K, Gourinath, S, Sharma, S, Ray, I, Gupta, M.N, Singh, T.P. | | Deposit date: | 2000-02-16 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhancement of enzyme activity through three-phase partitioning: crystal structure of a modified serine proteinase at 1.5 A resolution.
Protein Eng., 14, 2001
|
|
6Z54
 
 | | Crystal structure of CLK3 in complex with macrocycle ODS2003178 | | Descriptor: | 1,2-ETHANEDIOL, 11,15-Dimethyl-6-(oxan-4-yloxy)-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2003178
To Be Published
|
|
5CZL
 
 | | Crystal structure of a novel GH8 endo-beta-1,4-glucanase from an Achatina fulica gut metagenomic library | | Descriptor: | Glucanase, PHOSPHATE ION | | Authors: | Scapin, S.M.N, Souza, F.H.M, Zanphorlin, L.M, Almeida, T.S, Sade, Y.B, Cardoso, A.M, Pinheiro, G.L, Murakami, M.T. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Crystal structure of a novel GH8 endo-beta-1,4-glucanase from an Achatina fulica gut metagenomic library
To Be Published
|
|
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | Descriptor: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,Probable G-protein coupled receptor 34,YL-365 | | Authors: | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1EID
 
 | |
6WO7
 
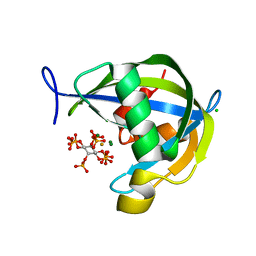 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5-Diphosphoinositol pentakisphosphate (5-IP7), Mg, and Fluoride ion | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
8Q6F
 
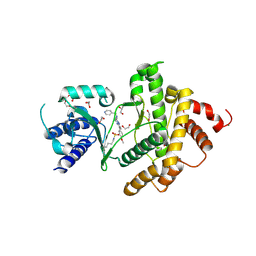 | | HUMAN PI4KIIIB IN COMPLEX WITH COVALENTLY BOUND INHIBITOR (COMPOUND 4) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-fluorosulfonyloxy-4-methoxy-phenyl)-2,5-dimethyl-7-(pyridin-4-ylmethylamino)pyrazolo[1,5-a]pyrimidine, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2023-08-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Covalent targeting of non-cysteine residues in PI4KIII beta.
Rsc Chem Biol, 4, 2023
|
|
6WX3
 
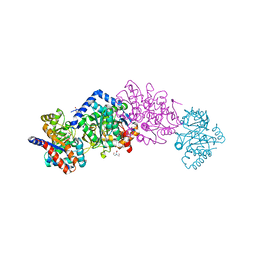 | | High resolution Tryptophan Synthase crystal structure from Salmonella typhimurium in complex with F9 inhibitor in the alpha-site, Cesium ion at the metal coordination site and internal aldimine form. | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, BICINE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution Tryptophan Synthase crystal structure from Salmonella typhimurium in complex with F9 inhibitor in the alpha-site, Cesium ion at the metal coordination site and internal aldimine form.
To be Published
|
|
8RX6
 
 | | Mycothione reductase from Mycobacterium tuberculosis in complex with Respiri-1093 | | Descriptor: | (3~{S})-4-[4-(1-benzothiophen-3-yl)-6,7,8,9-tetrahydro-5~{H}-pyrimido[4,5-d]azepin-2-yl]-3-methyl-morpholine, FLAVIN-ADENINE DINUCLEOTIDE, Mycothione reductase | | Authors: | Oorts, L, Osipov, E.M, Beelen, S, Strelkov, S.V. | | Deposit date: | 2024-02-06 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of mycothione reductase
To Be Published
|
|
8XH8
 
 | |
9HR6
 
 | |
6ZEJ
 
 | | Structure of PP1-Phactr1 chimera [PP1(7-304) + linker (SGSGS) + Phactr1(526-580)] | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
8XGE
 
 | | Human Cx36/GJD2 gap junction channel in porcine brain lipids. | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Cho, H.J, Lee, H.H. | | Deposit date: | 2023-12-15 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mefloquine-induced conformational shift in Cx36 N-terminal helix leading to channel closure mediated by lipid bilayer.
Nat Commun, 15, 2024
|
|
8XGF
 
 | | Human Cx36/GJD2 gap junction channel in complex with arachidonic acid. | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ARACHIDONIC ACID, Gap junction delta-2 protein | | Authors: | Cho, H.J, Lee, H.H. | | Deposit date: | 2023-12-15 | | Release date: | 2024-11-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Mefloquine-induced conformational shift in Cx36 N-terminal helix leading to channel closure mediated by lipid bilayer.
Nat Commun, 15, 2024
|
|
8XH9
 
 | |
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8J2W
 
 | | Saccharothrix syringae photocobilins protein, dark state | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-DEOXYADENOSINE, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
7S6S
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL1 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
7NT7
 
 | |
