6RS4
 
 | | Structure of tabersonine synthase - an alpha-beta hydrolase from Catharanthus roseus | | Descriptor: | 1,2-ETHANEDIOL, Tabersonine synthase | | Authors: | Caputi, L, Franke, J, Bussey, K, Farrow, S.C, Curcino Vieira, I.J, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2019-05-21 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids.
Nat.Chem.Biol., 16, 2020
|
|
6RT8
 
 | | Structure of catharanthine synthase - an alpha-beta hydrolase from Catharanthus roseus with a cleaviminium intermediate bound | | Descriptor: | 18-carboxymethoxy-cleaviminium, Catharanthine synthase, HEXAETHYLENE GLYCOL | | Authors: | Caputi, L, Franke, J, Bussey, K, Farrow, S.C, Curcino Vieira, I.J, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2019-05-22 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids.
Nat.Chem.Biol., 16, 2020
|
|
6RHL
 
 | | Room temperature data of Galectin-3C in complex with a pair of enantiomeric ligands: R enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{R})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Are crystallographic B-factors suitable for calculating protein conformational entropy?
Phys Chem Chem Phys, 21, 2019
|
|
6RHM
 
 | | Room temperature data of Galectin-3C in complex with a pair of enantiomeric ligands: S enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{S})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Are crystallographic B-factors suitable for calculating protein conformational entropy?
Phys Chem Chem Phys, 21, 2019
|
|
6RSQ
 
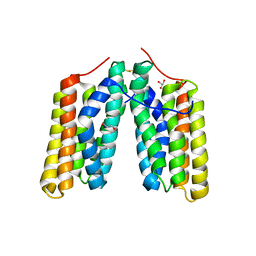 | | Helical folded domain of mouse CAP1 | | Descriptor: | Adenylyl cyclase-associated protein 1, GLYCEROL | | Authors: | Kotila, T, Kogan, K, Lappalainen, P. | | Deposit date: | 2019-05-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin.
Nat Commun, 10, 2019
|
|
6RZ2
 
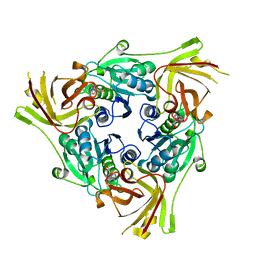 | | SalL with Chloroadenosine | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Adenosyl-chloride synthase | | Authors: | McKean, I, Frese, A, Cuetos, A, Burley, G, Grogan, G. | | Deposit date: | 2019-06-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4HPC
 
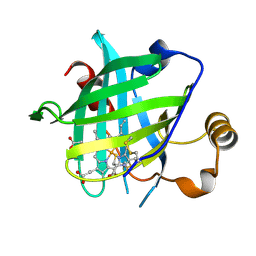 | |
4HPB
 
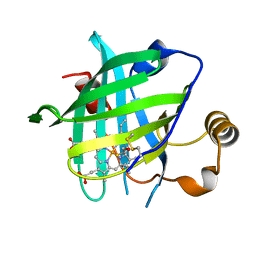 | |
6RZH
 
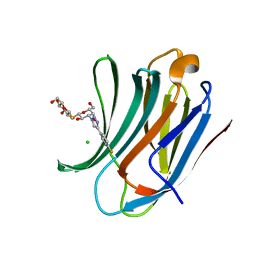 | | Galectin-3C in complex with para-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(4-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.947 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
4HPD
 
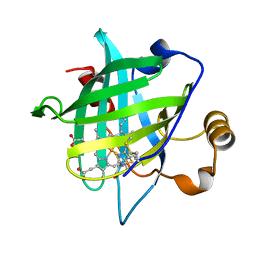 | |
4HPA
 
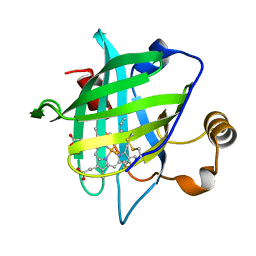 | |
6SF5
 
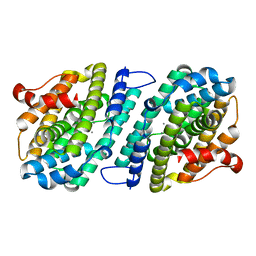 | | Mn-containing form of the ribonucleotide reductase NrdB protein from Leeuwenhoekiella blandensis | | Descriptor: | MANGANESE (II) ION, Ribonucleoside-diphosphate reductase, beta subunit 1 | | Authors: | Hasan, M, Rozman Grinberg, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2019-08-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Class Id ribonucleotide reductase utilizes a Mn2(IV,III) cofactor and undergoes large conformational changes on metal loading.
J.Biol.Inorg.Chem., 24, 2019
|
|
6SF4
 
 | | Apo form of the ribonucleotide reductase NrdB protein from Leeuwenhoekiella blandensis | | Descriptor: | Ribonucleoside-diphosphate reductase, beta subunit 1 | | Authors: | Hasan, M, Rozman Grinberg, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Class Id ribonucleotide reductase utilizes a Mn2(IV,III) cofactor and undergoes large conformational changes on metal loading.
J.Biol.Inorg.Chem., 24, 2019
|
|
6SLE
 
 | | Structure of Reductive Aminase from Neosartorya fumigata in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
6SUA
 
 | |
6SNU
 
 | | Crystal structure of the W60C mutant of the (S)-selective transaminase from Chromobacterium violaceum | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase family protein, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ruggieri, F, Gustafsson, C, Kimbung, R.Y, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-08-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Combined with Molecular Dynamics Reveal Altered Flow of Water in the Active Site of W60C Chromobacterium violaceum omega-transaminase
Not Published
|
|
6T3B
 
 | | Crystal structure of PI3Kgamma with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Oster, L, Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6SKX
 
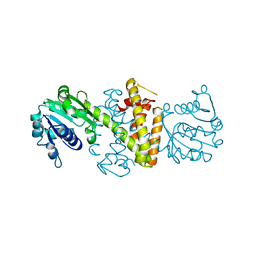 | | Structure of Reductive Aminase from Neosartorya fumigata | | Descriptor: | Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
6RA4
 
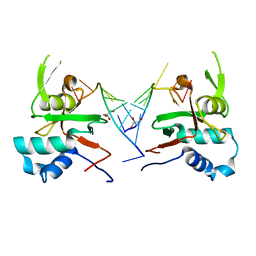 | | Human ARGONAUTE-2 PAZ DOMAIN (214-347) IN COMPLEX WITH CGUGACUCU | | Descriptor: | GLYCEROL, Protein argonaute-2, RNA (5'-R(*CP*GP*UP*GP*AP*CP*UP*CP*U)-3') | | Authors: | Rondeau, J.-M, Bourgier, E. | | Deposit date: | 2019-04-05 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How to Computationally Stack the Deck for Hit-to-Lead Generation: In Silico Molecular Interaction Energy Profiling for de Novo siRNA Guide Strand Surrogate Selection.
J.Chem.Inf.Model., 59, 2019
|
|
6TF7
 
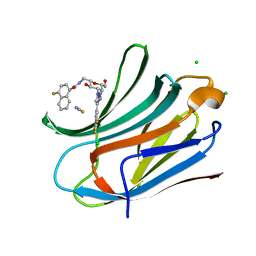 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | 4-fluoranyl-~{N}-[[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-2-yl]methyl]naphthalene-1-carboxamide, CHLORIDE ION, Galectin-3, ... | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
4P3S
 
 | |
4P3T
 
 | |
6RYZ
 
 | | SalL with S-adenosyl methionine | | Descriptor: | 1,2-ETHANEDIOL, Adenosyl-chloride synthase, CHLORIDE ION, ... | | Authors: | McKean, I, Frese, A, Cuetos, A, Burley, G, Grogan, G. | | Deposit date: | 2019-06-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6RZF
 
 | | Galectin-3C in complex with ortho-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(2-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.016 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
