2KJ1
 
 | |
1WP5
 
 | |
2KLM
 
 | | Solution Structure of L11 with SAXS and RDC | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Wang, J, Zuo, X, Yu, P, Schwieters, C.D, Wang, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
2KM0
 
 | | Cu(I)-bound CopK | | Descriptor: | COPPER (I) ION, Copper resistance protein K | | Authors: | Bersch, B. | | Deposit date: | 2009-07-15 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CopK from Cupriavidus metallidurans CH34 Binds Cu(I) in a Tetrathioether Site: Characterization by X-ray Absorption and NMR Spectroscopy
J.Am.Chem.Soc., 2010
|
|
4RA8
 
 | | Structure analysis of the Mip1a P8A mutant | | Descriptor: | C-C motif chemokine 3 | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RAL
 
 | | Crystal structure of insulin degrading enzyme in complex with macrophage inflammatory protein 1 beta | | Descriptor: | C-C motif chemokine 4, Insulin-degrading enzyme, ZINC ION | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
7PWI
 
 | | Structure of the dTDP-sugar epimerase StrM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, dTDP-4-keto-rhamnose 3,5-epimerase,dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.326 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
1N83
 
 | | Crystal Structure of the complex between the Orphan Nuclear Hormone Receptor ROR(alpha)-LBD and Cholesterol | | Descriptor: | CHOLESTEROL, Nuclear receptor ROR-alpha | | Authors: | Kallen, J.A, Schlaeppi, J.M, Bitsch, F, Geisse, S, Geiser, M, Delhon, I, Fournier, B. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Structure of hROR(alpha) LBD at 1.63A: Structural and Functional data that Cholesterol or a Cholesterol derivative is the natural ligand of ROR(alpha)
Structure, 10, 2002
|
|
5HY2
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
2GI7
 
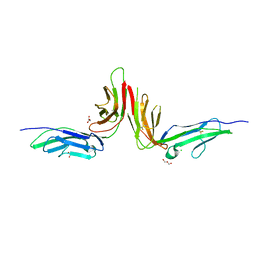 | | Crystal structure of human platelet Glycoprotein VI (GPVI) | | Descriptor: | CHLORIDE ION, GLYCEROL, GPVI protein, ... | | Authors: | Horii, K, Kahn, M.L, Herr, A.B. | | Deposit date: | 2006-03-28 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for platelet collagen responses by the immune-type receptor glycoprotein VI.
Blood, 108, 2006
|
|
5HHM
 
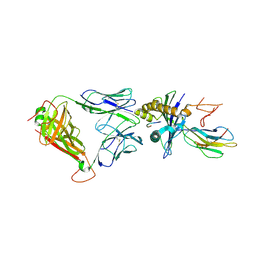 | | Crystal Structure of the JM22 TCR in complex with HLA-A*0201 in complex with M1-F5L | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Gras, S, Josephs, T.M, Rossjohn, J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5O3B
 
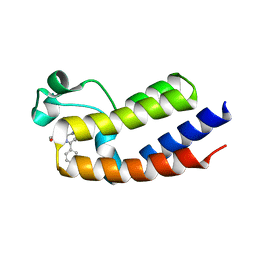 | | Human Brd2(BD2) mutant in complex with AL | | Descriptor: | Bromodomain-containing protein 2, methyl (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]pent-4-enoate | | Authors: | Runcie, A.C, Chan, K.-H, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5A5B
 
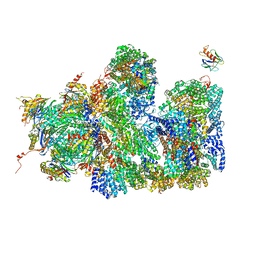 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
9F5T
 
 | | Ubiquitin C-terminal clippase BpJOS | | Descriptor: | BpJOS, Ubiquitin | | Authors: | Uthoff, M, Hermanns, T, Birkmann, S, Hofmann, K, Baumann, U. | | Deposit date: | 2024-04-30 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A family of bacterial Josephin-like deubiquitinases with an irreversible cleavage mode.
Mol.Cell, 85, 2025
|
|
9FPA
 
 | |
5HWE
 
 | | high resolution structure of barbiturase | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, MAGNESIUM ION, ... | | Authors: | Peat, T.S, Scott, C. | | Deposit date: | 2016-01-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5O3H
 
 | | Human Brd2(BD2) mutant in complex with 9-ME-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-9-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-propanamide, (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5HVP
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF A COMPLEX BETWEEN HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE AND ACETYL-PEPSTATIN AT 2.0-ANGSTROMS RESOLUTION | | Descriptor: | ACETYL-*PEPSTATIN, CHLORIDE ION, HIV-1 PROTEASE | | Authors: | Fitzgerald, P.M.D, Mckeever, B.M, Vanmiddlesworth, J.F, Springer, J.P. | | Deposit date: | 1990-04-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of a complex between human immunodeficiency virus type 1 protease and acetyl-pepstatin at 2.0-A resolution.
J.Biol.Chem., 265, 1990
|
|
1WVT
 
 | | Crystal structure of uncharacterized protein ST2180 from Sulfolobus tokodaii | | Descriptor: | hypothetical protein ST2180 | | Authors: | Sasaki, T, Tanaka, Y, Yasutake, Y, Tsumoto, K, Tanaka, I, Kumagai, I. | | Deposit date: | 2004-12-27 | | Release date: | 2006-01-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular properties of two proteins homologous to PduO-type ATP:cob(I)alamin adenosyltransferase from Sulfolobus tokodaii
Proteins, 68, 2007
|
|
5HXZ
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
2LCT
 
 | |
2LCC
 
 | | Solution structure of RBBP1 chromobarrel domain | | Descriptor: | AT-rich interactive domain-containing protein 4A | | Authors: | Gong, W, Feng, Y. | | Deposit date: | 2011-04-28 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into recognition of methylated histone tails by retinoblastoma-binding protein 1.
J.Biol.Chem., 2012
|
|
5O3G
 
 | | Human Brd2(BD2) mutant in complex with AL-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-pent-4-enamide, Bromodomain-containing protein 2 | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
2RVQ
 
 | | Solution structure of the isolated histone H2A-H2B heterodimer | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J | | Authors: | Moriwaki, Y, Yamane, T, Ohtomo, H, Ikeguchi, M, Kurita, J, Sato, M, Nagadoi, A, Shimojo, H, Nishimura, Y. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the isolated histone H2A-H2B heterodimer
Sci Rep, 6, 2016
|
|
5OCZ
 
 | | Free DNA_hairpin polyamides studies | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*G)-3') | | Authors: | Padroni, G, Parkinson, J, Burley, G.A. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides.
Nucleic Acids Res., 46, 2018
|
|
