1U1J
 
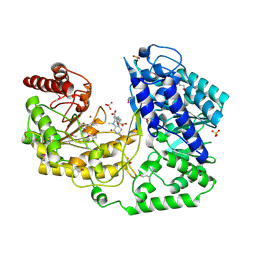 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, METHIONINE, ... | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-15 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
2DG3
 
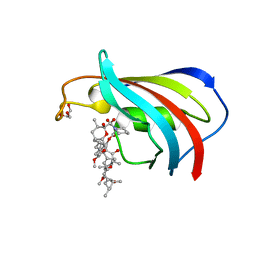 | | Wildtype FK506-binding protein complexed with Rapamycin | | Descriptor: | FK506-binding protein 1A, GLYCEROL, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Buckle, A.M. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Energetic and structural analysis of the role of tryptophan 59 in FKBP12
Biochemistry, 42, 2003
|
|
2DGB
 
 | |
6F0A
 
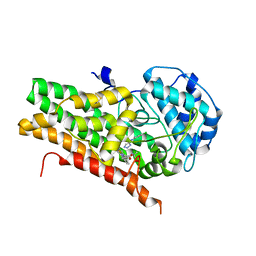 | | Crystal structure of human indoleamine 2,3-dioxygenase bound to a triazole inhibitor and alanine molecule. | | Descriptor: | ALANINE, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Swan, M.K, Latchem, M. | | Deposit date: | 2017-11-17 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | New 4-Amino-1,2,3-Triazole Inhibitors of Indoleamine 2,3-Dioxygenase Form a Long-Lived Complex with the Enzyme and Display Exquisite Cellular Potency.
Chembiochem, 19, 2018
|
|
1U11
 
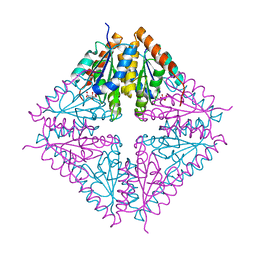 | | PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) from the acidophile Acetobacter aceti | | Descriptor: | CITRIC ACID, PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) | | Authors: | Settembre, E.C, Chittuluru, J.R, Mill, C.P, Kappock, T.J, Ealick, S.E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acidophilic adaptations in the structure of Acetobacter aceti N5-carboxyaminoimidazole ribonucleotide mutase (PurE).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U1U
 
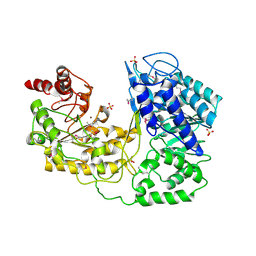 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, SULFATE ION, ZINC ION | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
2DWC
 
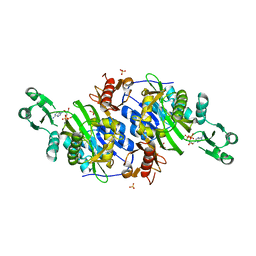 | | Crystal structure of Probable phosphoribosylglycinamide formyl transferase from Pyrococcus horikoshii OT3 complexed with ADP | | Descriptor: | 433aa long hypothetical phosphoribosylglycinamide formyl transferase, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Probable phosphoribosylglycinamide formyl transferase from Pyrococcus horikoshii OT3 complexed with ADP
To be Published
|
|
6ZWZ
 
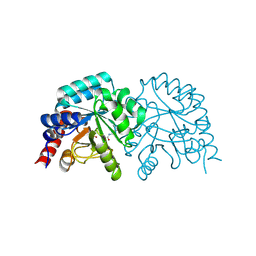 | |
6ZX1
 
 | | OMPD-domain of human UMPS in complex with 6-Aza-UMP at 1.0 Angstroms resolution | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, PROLINE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX0
 
 | | OMPD-domain of human UMPS in complex with the substrate OMP at 1.25 Angstroms resolution | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZWY
 
 | |
6ZX3
 
 | | OMPD-domain of human UMPS in complex with 6-thiocarboxamido-UMP at 1.15 Angstroms resolution | | Descriptor: | GLYCEROL, PROLINE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Schimdt, T. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX2
 
 | | OMPD-domain of human UMPS in complex with 6-carboxamido-UMP at 1.2 Angstroms resolution | | Descriptor: | PROLINE, SULFATE ION, Uridine 5'-monophosphate synthase, ... | | Authors: | Tittmann, K, Rindfleisch, S, Schimdt, T. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7ODX
 
 | |
3AJX
 
 | | Crystal Structure of 3-Hexulose-6-Phosphate Synthase | | Descriptor: | 3-hexulose-6-phosphate synthase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kita, A, Orita, I, Yurimoto, H, Kato, N, Sakai, Y, Miki, K. | | Deposit date: | 2010-06-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of 3-hexulose-6-phosphate synthase, a member of the orotidine 5'-monophosphate decarboxylase suprafamily
Proteins, 78, 2010
|
|
3AV3
 
 | | Crystal structure of glycinamide ribonucleotide transformylase 1 from Geobacillus kaustophilus | | Descriptor: | MAGNESIUM ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Kanagawa, M, Baba, S, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-02-18 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
5EA9
 
 | | Crystal Structure of Trypanosoma cruzi Dihydroorotate Dehydrogenase in Complex with Neq0130 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-3-thiophen-2-ylprop-2-enylidene]-1,3-diazinane-2,4,6-trione, COBALT HEXAMMINE(III), ... | | Authors: | Rocha, J.R, Inaoka, D.K, Cheleski, J, Shiba, T, Harada, S, Montanari, C.A, Kita, K. | | Deposit date: | 2015-10-15 | | Release date: | 2016-10-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Exploring Trypanosoma cruzi Dihydroorotate Dehydrogenase Active Site Plasticity for the Discovery of Potent and Selective Inhibitors with Trypanocidal Activity
To be Published
|
|
5E93
 
 | | Crystal Structure of Trypanosoma cruzi Dihydroorotate Dehydrogenase in Complex with Neq0071 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-3-(furan-2-yl)prop-2-enylidene]-1,3-diazinane-2,4,6-trione, CACODYLATE ION, ... | | Authors: | Rocha, J.R, Inaoka, D.K, Cheleski, J, Shiba, T, Harada, S, Montanari, C.A, Kita, K. | | Deposit date: | 2015-10-14 | | Release date: | 2016-10-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Exploring Trypanosoma cruzi Dihydroorotate Dehydrogenase Active Site Plasticity for the Discovery of Potent and Selective Inhibitors with Trypanocidal Activity
To be Published
|
|
5OWI
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 1) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
6LKD
 
 | | in meso full-length rat KMO in complex with a pyrazoyl benzoic acid inhibitor | | Descriptor: | 5-[5-(4-chloranyl-3-fluoranyl-phenyl)-4-methyl-pyrazol-1-yl]-2-phenylmethoxy-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
5OWJ
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 2) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
6LKE
 
 | | in meso full-length rat KMO in complex with an inhibitor identified via DNA-encoded chemical library screening | | Descriptor: | 4-chloranyl-2-[[5-chloranyl-2-(5-methoxy-1,3-dihydroisoindol-2-yl)-1,3-thiazol-4-yl]carbonyl-methyl-amino]-5-fluoranyl-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Hupp, D.C, Liu, J, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
6OTT
 
 | | Structure of PurF in complex with ppApp | | Descriptor: | Amidophosphoribosyltransferase, PurF, MAGNESIUM ION, ... | | Authors: | Grant, R.A, Wang, B, Laub, M.T. | | Deposit date: | 2019-05-03 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp.
Nature, 575, 2019
|
|
4MA0
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP
To be Published
|
|
4MAM
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP
To be Published
|
|
