8K7F
 
 | | Crystal structure of human lysosomal alpha-galactosidase A in complex with (2R,3S,4R,5R)-2,5-bis(hydroxymethyl)pyrrolidine-3,4-diol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2,5-bis(hydroxymethyl)pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Y, Huang, K.F, Ko, T.P, Cheng, W.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human alpha-Galactosidase.
Jacs Au, 4, 2024
|
|
8K7E
 
 | | Crystal structure of human lysosomal alpha-galactosidase A in complex with (2R,3R,4S,5R)-2-(aminomethyl)-5-(hydroxymethyl)pyrrolidine-3,4-diol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(aminomethyl)-5-(hydroxymethyl)pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Y, Huang, K.F, Ko, T.P, Cheng, W.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human alpha-Galactosidase.
Jacs Au, 4, 2024
|
|
8K7K
 
 | | Crystal structure of human lysosomal alpha-galactosidase A in complex with (2R,3S,4R,5R)-2,5-bis(hydroxymethyl)-1-methylpyrrolidine-3,4-diol | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2,5-bis(hydroxymethyl)-1-methyl-pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Y, Huang, K.F, Ko, T.P, Cheng, W.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human alpha-Galactosidase.
Jacs Au, 4, 2024
|
|
1H5F
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (22-33% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
3ML2
 
 | | Human carbonic anhydsase II in complex with an aryl sulfonamide inhibitor | | Descriptor: | 2-(7-methoxy-2-oxo-2H-chromen-4-yl)-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, Mckenna, R. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coumarinyl-substituted sulfonamides strongly inhibit several human carbonic anhydrase isoforms: solution and crystallographic investigations.
Bioorg.Med.Chem., 18, 2010
|
|
1CDE
 
 | | STRUCTURES OF APO AND COMPLEXED ESCHERICHIA COLI GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | 5-DEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Almassy, R.J, Janson, C.A, Kan, C.-C, Hostomska, Z. | | Deposit date: | 1992-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1QXK
 
 | | Monoacid-Based, Cell Permeable, Selective Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | 2-{4-[2-ACETYLAMINO-3-(4-CARBOXYMETHOXY-3-HYDROXY-PHENYL)-PROPIONYLAMINO]-BUTOXY}-6-HYDROXY-BENZOIC ACID METHYL ESTER, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Xin, Z, Liu, G, Abad-Zapatero, C, Pei, Z, Szczepankiewick, B.G, Li, X, Zhang, T, Hutchins, C.W, Hajduk, P.J, Ballaron, S.J, Stashko, M.A, Lubben, T.H, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-09-08 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a Monoacid-Based, Cell Permeable, Selective
Inhibitor of Protein Tyrosine Phosphatase 1B
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
8JNW
 
 | | PfDXR - Mn2+ - NADPH - MAMK89 quaternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Reverse N -Substituted Hydroxamic Acid Derivatives of Fosmidomycin Target a Previously Unknown Subpocket of 1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase (DXR).
Acs Infect Dis., 10, 2024
|
|
8HKW
 
 | |
8JNV
 
 | | PfDXR - Mn2+ - MAMK89 ternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2023-06-06 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reverse N -Substituted Hydroxamic Acid Derivatives of Fosmidomycin Target a Previously Unknown Subpocket of 1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase (DXR).
Acs Infect Dis., 10, 2024
|
|
8HIK
 
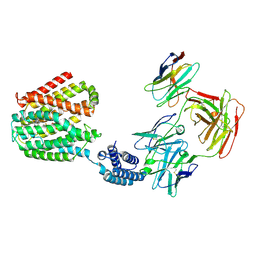 | | The TPP-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HII
 
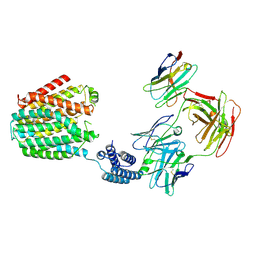 | | The BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | BRIL-SLC19A1 chimera, anti-BRIL Fab heavy chain, anti-BRIL Fab light chain, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
2ING
 
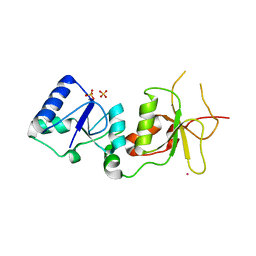 | | X-ray Structure of the BRCA1 BRCT mutant M1775K | | Descriptor: | Breast cancer type 1 susceptibility protein, COBALT (II) ION, SULFATE ION | | Authors: | Birrane, G, Soni, A, Ladias, J.A.A. | | Deposit date: | 2006-10-07 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Pathogenicity of the BRCA1 missense variant M1775K is determined by the disruption of the BRCT phosphopeptide-binding pocket: a multi-modal approach.
Eur.J.Hum.Genet., 16, 2008
|
|
3AZM
 
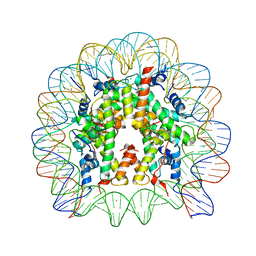 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K79Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZL
 
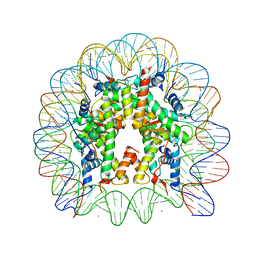 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K77Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
1XOY
 
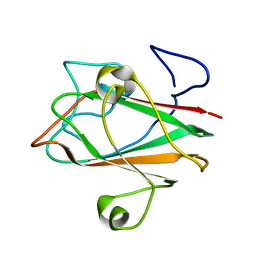 | | Solution structure of At3g04780.1, an Arabidopsis ortholog of the C-terminal domain of human thioredoxin-like protein | | Descriptor: | hypothetical protein At3g04780.1 | | Authors: | Song, J, Robert, C.T, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of At3g04780.1-des15, an Arabidopsis thaliana ortholog of the C-terminal domain of human thioredoxin-like protein.
Protein Sci., 14, 2005
|
|
1QTY
 
 | |
1XO8
 
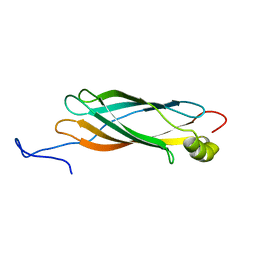 | | Solution structure of AT1g01470 from Arabidopsis Thaliana | | Descriptor: | At1g01470 | | Authors: | Singh, S, Cornilescu, C.C, Tyler, R.C, Cornilescu, G, Tonelli, M, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-06 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein
Protein Sci., 14, 2005
|
|
3OIK
 
 | | Human Carbonic anhydrase II mutant A65S, N67Q (CA IX mimic) bound by 2-Ethylestradiol 3,17-O,O-bis-sulfamate | | Descriptor: | (9BETA,13ALPHA,14BETA,17ALPHA)-2-ETHYLESTRA-1(10),2,4-TRIENE-3,17-DIYL DISULFAMATE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
2IX3
 
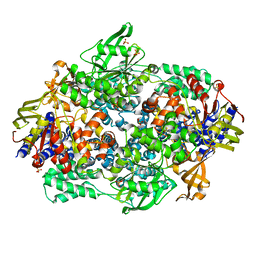 | | Structure of yeast Elongation Factor 3 | | Descriptor: | ELONGATION FACTOR 3, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
3AZN
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K91Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZF
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K79Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
1H9H
 
 | | COMPLEX OF EETI-II WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Wentzel, A, Kolmar, H, Uson, I. | | Deposit date: | 2001-03-12 | | Release date: | 2004-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6ZE6
 
 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment 4-nitrocatechol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITROCATECHOL, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8GYH
 
 | | Crystal structure of Fic25 (apo form) from Streptomyces ficellus | | Descriptor: | DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, IMIDAZOLE | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
