8FEN
 
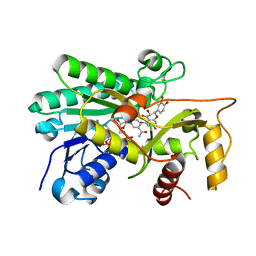 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP and DHQ | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Panicum virgatum Dihydroflavonol 4-Reductase | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
3IUB
 
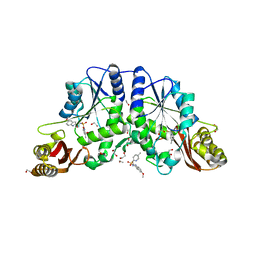 | | Crystal structure of pantothenate synthetase from Mycobacterium tuberculosis in complex with 5-Methoxy-N-(5-methylpyridin-2-ylsulfonyl)-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-methoxy-N-[(5-methylpyridin-2-yl)sulfonyl]-1H-indole-2-carboxamide, ETHANOL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3EGO
 
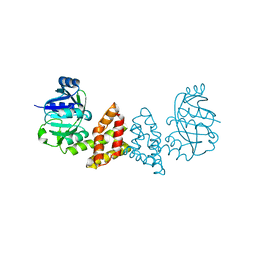 | | Crystal structure of Probable 2-dehydropantoate 2-reductase panE from Bacillus Subtilis | | Descriptor: | Probable 2-dehydropantoate 2-reductase | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Hu, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-11 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Probable 2-dehydropantoate 2-reductase panE from Bacillus Subtilis
To be published
|
|
8EST
 
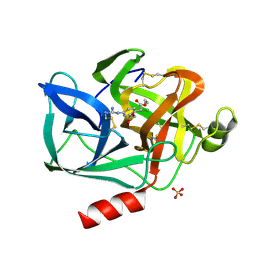 | | REACTION OF PORCINE PANCREATIC ELASTASE WITH 7-SUBSTITUTED 3-ALKOXY-4-CHLOROISOCOUMARINS: DESIGN OF POTENT INHIBITORS USING THE CRYSTAL STRUCTURE OF THE COMPLEX FORMED WITH 4-CHLORO-3-ETHOXY-7-GUANIDINO-ISOCOUMARIN | | Descriptor: | CALCIUM ION, ETHYL-(2-CARBOXY-4-GUANIDINIUM-PHENYL)-CHLOROACETATE, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Radhakrishnan, R, Powers, J.C, Meyerjunior, E.F. | | Deposit date: | 1990-02-21 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Reaction of porcine pancreatic elastase with 7-substituted 3-alkoxy-4-chloroisocoumarins: design of potent inhibitors using the crystal structure of the complex formed with 4-chloro-3-ethoxy-7-guanidinoisocoumarin.
Biochemistry, 29, 1990
|
|
1K6E
 
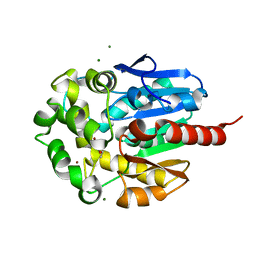 | | COMPLEX OF HYDROLYTIC HALOALKANE DEHALOGENASE LINB FROM SPHINGOMONAS PAUCIMOBILIS UT26 WITH 1,2-PROPANEDIOL (PRODUCT OF DEHALOGENATION OF 1,2-DIBROMOPROPANE) AT 1.85A | | Descriptor: | 1-BROMOPROPANE-2-OL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Streltsov, V.A, Prokop, Z, Damborsky, J, Nagata, Y, Oakley, A, Wilce, M.C.J. | | Deposit date: | 2001-10-16 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates.
Biochemistry, 42, 2003
|
|
1QR0
 
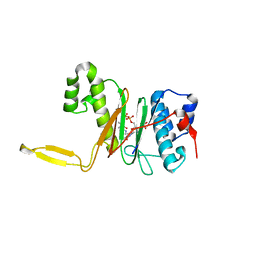 | | CRYSTAL STRUCTURE OF THE 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP-COENZYME A COMPLEX | | Descriptor: | 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP, COENZYME A, MAGNESIUM ION | | Authors: | Reuter, K, Mofid, R.M, Marahiel, A.M, Ficner, R. | | Deposit date: | 1999-06-17 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily.
EMBO J., 18, 1999
|
|
1F2J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ALDOLASE FROM T. BRUCEI | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE, GLYCOSOMAL | | Authors: | Chudzik, D.M, Michels, P.A, De Walque, S, Hol, W.G.J. | | Deposit date: | 2000-05-25 | | Release date: | 2000-07-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of type 2 peroxisomal targeting signals in two trypanosomatid aldolases.
J.Mol.Biol., 300, 2000
|
|
1OU6
 
 | |
5YG2
 
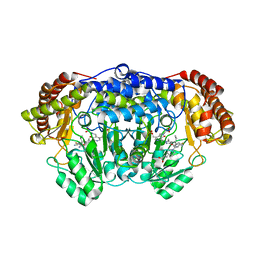 | | Plasmodium vivax SHMT bound with PLP-glycine and GS705 | | Descriptor: | 3-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]piperidin-4-yl]propanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
1KPG
 
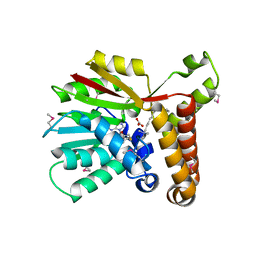 | | Crystal Structure of mycolic acid cyclopropane synthase CmaA1 complexed with SAH and CTAB | | Descriptor: | CARBONATE ION, CETYL-TRIMETHYL-AMMONIUM, CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 1, ... | | Authors: | Huang, C.-C, Smith, C.V, Jacobs Jr, W.R, Glickman, M.S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-30 | | Release date: | 2002-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis
J.Biol.Chem., 277, 2002
|
|
3F3M
 
 | |
4CRZ
 
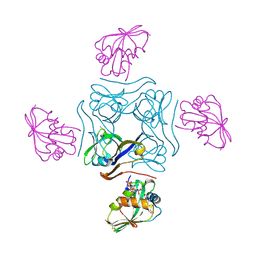 | | Direct visualisation of strain-induced protein prost-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Pand/Panz Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A.
Chem.Biol., 22, 2015
|
|
4CS0
 
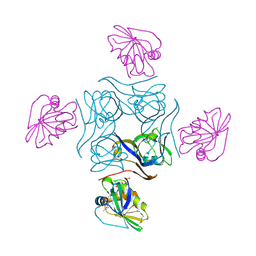 | | Direct visualisation of strain-induced protein post-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Pand/Panz Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A.
Chem.Biol., 22, 2015
|
|
3JW1
 
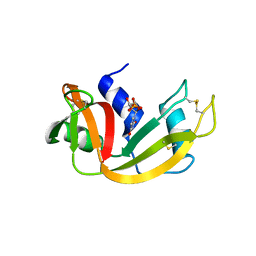 | | Crystal Structure of Bovine Pancreatic Ribonuclease Complexed with Uridine-5'-monophosphate at 1.60 A Resolution | | Descriptor: | Ribonuclease pancreatic, URIDINE-5'-MONOPHOSPHATE | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, Mcpherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic ribonuclease complexed with uridine 5'-monophosphate at 1.60 A resolution.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7FTD
 
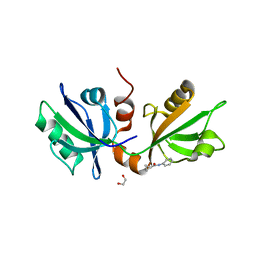 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z27782662 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
3KSL
 
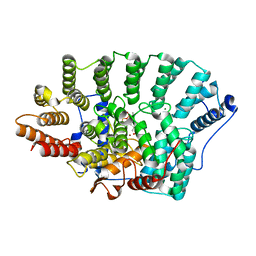 | | Structure of FPT bound to DATFP-DH-GPP | | Descriptor: | (2S,6E)-8-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}-2,6-dimethyloct-6-en-1-yl (2S)-3,3,3-trifluoro-2-hydrazinopropanoate, Farnesyltransferase, CAAX box, ... | | Authors: | Hovlid, M.L, Edelstein, R.L, Henry, O, Ochocki, J, DeGraw, A, Lenevich, S, Talbot, T, Young, V, Hruza, A.W, Lopez-Gallego, F, Labello, N.P, Strickland, C.L, Schmidt-Dannert, C, Distefano, M.D. | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis, properties, and applications of diazotrifluropropanoyl-containing photoactive analogs of farnesyl diphosphate containing modified linkages for enhanced stability.
Chem.Biol.Drug Des., 75, 2010
|
|
3L2L
 
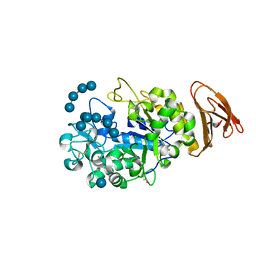 | | X-ray Crystallographic Analysis of Pig Pancreatic Alpha-Amylase with Limit Dextrin and Oligosaccharide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-ray crystallographic analyses of pig pancreatic alpha-amylase with limit dextrin, oligosaccharide, and alpha-cyclodextrin.
Biochemistry, 49, 2010
|
|
3L2M
 
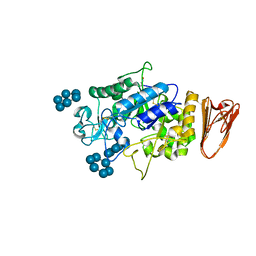 | | X-ray Crystallographic Analysis of Pig Pancreatic Alpha-Amylase with Alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | X-ray crystallographic analyses of pig pancreatic alpha-amylase with limit dextrin, oligosaccharide, and alpha-cyclodextrin.
Biochemistry, 49, 2010
|
|
6AKY
 
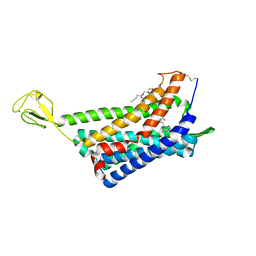 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 34 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-3-yl)propyl]cyclohexane-1-carboxamide, C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ... | | Authors: | Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
2F8W
 
 | | Crystal structure of d(CACGTG)2 | | Descriptor: | 1,3-DIAMINOPROPANE, 5'-D(*CP*AP*CP*GP*TP*G)-3', SPERMINE | | Authors: | Narayana, N, Shamala, N, Ganesh, K.N, Viswamitra, M.A. | | Deposit date: | 2005-12-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interaction between the Z-Type DNA Duplex and 1,3-Propanediamine: Crystal Structure of d(CACGTG)2 at 1.2 A Resolution
Biochemistry, 45, 2006
|
|
3IJ8
 
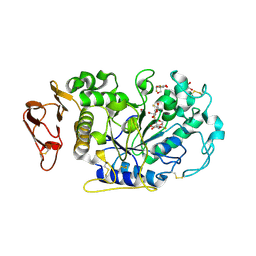 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, 5-fluoro-alpha-L-idopyranose, CALCIUM ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
5XMR
 
 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and GS395 | | Descriptor: | (4~{S})-6-azanyl-3-methyl-4-[3-[4-(phenylmethyl)sulfonylphenyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-2~{H}-pyrano[2 ,3-c]pyrazole-5-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
3IJ7
 
 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | 4-O-methyl-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranosyl fluoride, 4-O-methyl-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-2)-5-fluoro-alpha-L-idopyranose, CALCIUM ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-03 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
3IJ9
 
 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
5X7D
 
 | | Structure of beta2 adrenoceptor bound to carazolol and an intracellular allosteric antagonist | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liu, X, Ahn, S, Kahsai, A.W, Meng, K.-C, Latorraca, N.R, Pani, B, Venkatakrishnan, A.J, Masoudi, A, Weis, W.I, Dror, R.O, Chen, X, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2017-02-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Mechanism of intracellular allosteric beta 2AR antagonist revealed by X-ray crystal structure.
Nature, 548, 2017
|
|
