3P60
 
 | | Crystal structure of the mutant T159V of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
7ETG
 
 | | Crystal structure of AbHpaI-Co-pyruvate-succinic semialdehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, 4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETH
 
 | | Crystal structure of AbHpaI-Zn-pyruvate-propionaldehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, PROPANAL, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETD
 
 | | Crystal structure of AbHpaI-Zn-(4S)-KDGlu complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETE
 
 | | Crystal structure of AbHpaI-Mg-(4R)-KDGal complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | (4R,5R)-4,5,6-tris(oxidanyl)-2-oxidanylidene-hexanoic acid, 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7EQX
 
 | | Crystal structure of an Aedes aegypti procarboxypeptidase B1 | | Descriptor: | Carboxypeptidase B, ZINC ION | | Authors: | Choong, Y.K, Gavor, E, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-05-05 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of Aedes aegypti procarboxypeptidase B1 and its binding with Dengue virus for controlling infection.
Life Sci Alliance, 5, 2022
|
|
7ETI
 
 | | Crystal structure of AbHpaI-Zn-pyruvate-4-hydroxybenzaldehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, P-HYDROXYBENZALDEHYDE, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7FBP
 
 | | FXIIa-cMCoFx1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIIa light chain, cMCoFx1 | | Authors: | Sengoku, T, Liu, W, de Veer, S.J, Huang, Y.H, Okada, C, Zdenek, C.N, Fry, B.G, Swedberg, J.E, Passioura, T, Craik, D.J, Suga, H, Ogata, K. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An Ultrapotent and Selective Cyclic Peptide Inhibitor of Human beta-Factor XIIa in a Cyclotide Scaffold.
J.Am.Chem.Soc., 143, 2021
|
|
7EWP
 
 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:1:1 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
4HIA
 
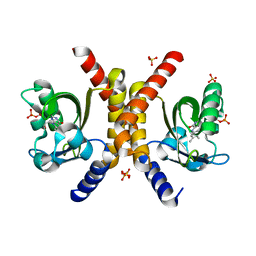 | |
7EWL
 
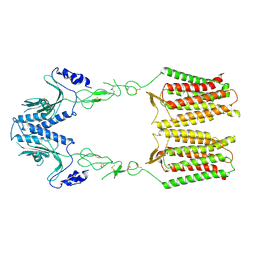 | | cryo-EM structure of apo GPR158 | | Descriptor: | Probable G-protein coupled receptor 158 | | Authors: | Jeong, E, Kim, Y, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
7EWR
 
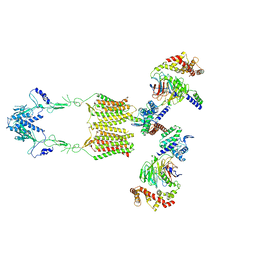 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:2:2 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
4HNU
 
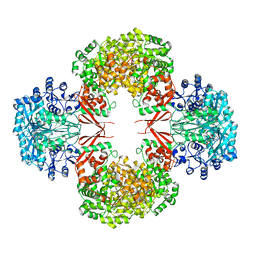 | | crystal structure of K442E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
4HNV
 
 | | Crystal structure of R54E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
1UXL
 
 | | I113T mutant of human SOD1 | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Hough, M.A, Grossmann, J.G, Antonyuk, S.V, Strange, R.W, Doucette, P.A, Rodriguez, J.A, Whitson, L.J, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2004-02-25 | | Release date: | 2004-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimer Destabilization in Superoxide Dismutase May Result in Disease-Causing Properties: Structures of Motor Neuron Disease Mutants
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4HJ4
 
 | |
3OF1
 
 | | Crystal Structure of Bcy1, the Yeast Regulatory Subunit of PKA | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase regulatory subunit | | Authors: | Rinaldi, J, Wu, J, Yang, J, Ralston, C.Y, Sankaran, B, Moreno, S, Taylor, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of Yeast Regulatory Subunit: A Glimpse into the Evolution of PKA Signaling.
Structure, 18, 2010
|
|
3ODR
 
 | |
1UYV
 
 | |
1UYR
 
 | |
1UB3
 
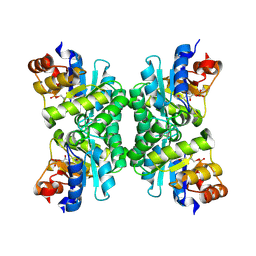 | | Crystal Structure of Tetrameric Structure of Aldolase from thermus thermophilus HB8 | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, Aldolase protein | | Authors: | Lokanath, N.K, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-28 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3G6W
 
 | |
1UYS
 
 | |
3SJ3
 
 | | Crystal structure of the mutant R160A.Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
4HNT
 
 | | crystal structure of F403A mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
