3D2G
 
 | |
2WTE
 
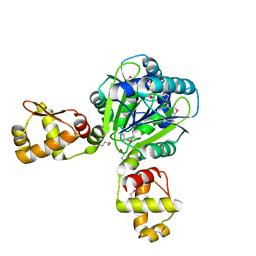 | | The structure of the CRISPR-associated protein, Csa3, from Sulfolobus solfataricus at 1.8 angstrom resolution. | | Descriptor: | CSA3, DI(HYDROXYETHYL)ETHER | | Authors: | Lintner, N.G, Alsbury, D.L, Copie, V, Young, M.J, Lawrence, C.M. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Crispr-Associated Protein Csa3 Provides Insight Into the Regulation of the Crispr/Cas System.
J.Mol.Biol., 405, 2011
|
|
7EDL
 
 | |
7EDM
 
 | |
7EDN
 
 | |
1SC7
 
 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Indenoisoquinoline MJ-II-38 and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 4-(5,11-DIOXO-5H-INDENO[1,2-C]ISOQUINOLIN-6(11H)-YL)BUTANOATE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-02-11 | | Release date: | 2005-04-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
1SEU
 
 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Indolocarbazole SA315F and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 2,10-DIHYDROXY-12-(BETA-D-GLUCOPYRANOSYL)-6,7,12,13-TETRAHYDROINDOLO[2,3-A]PYRROLO[3,4-C]CARBAZOLE-5,7-DIONE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-02-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
2MXD
 
 | | Solution structure of VPg of porcine sapovirus | | Descriptor: | Viral protein genome-linked | | Authors: | Kim, J, Hwang, H, Min, H, Yun, H, Cho, K, Pelton, J.G, Wemmer, D.E, Lee, C. | | Deposit date: | 2014-12-24 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the porcine sapovirus VPg core reveals a stable three-helical bundle with a conserved surface patch.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
5F9S
 
 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 1.7 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
3MWL
 
 | | Q28E mutant of HERA N-terminal RecA-like domain in complex with 8-OXOADENOSINE | | Descriptor: | 6-azanyl-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-7H-purin-8-one, Heat resistant RNA dependent ATPase, SULFATE ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
3MWJ
 
 | |
1R2Z
 
 | | MutM (Fpg) bound to 5,6-dihydrouracil (DHU) containing DNA | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(DHU)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | DNA Lesion Recognition by the Bacterial Repair Enzyme MutM.
J.Biol.Chem., 278, 2003
|
|
5JMV
 
 | | Crystal structure of mjKae1-pfuPcc1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Probable bifunctional tRNA threonylcarbamoyladenosine biosynthesis protein, ... | | Authors: | Wan, L, Sicheri, F. | | Deposit date: | 2016-04-29 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3864696 Å) | | Cite: | Structural and functional characterization of KEOPS dimerization by Pcc1 and its role in t6A biosynthesis.
Nucleic Acids Res., 44, 2016
|
|
1R2Y
 
 | | MutM (Fpg) bound to 8-oxoguanine (oxoG) containing DNA | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | DNA Lesion Recognition by the Bacterial Repair Enzyme MutM.
J.Biol.Chem., 278, 2003
|
|
2XC6
 
 | |
2X2Q
 
 | | Crystal structure of an 'all locked' LNA duplex at 1.9 angstrom resolution | | Descriptor: | CACODYLATE ION, COBALT HEXAMMINE(III), LOCKED NUCLEIC ACID DERIVED FROM TRNA SER ACCEPTOR STEM MICROHELIX, ... | | Authors: | Eichert, A, Behling, K, Fuerste, J.P, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2010-01-15 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of an 'All Locked' Nucleic Acid Duplex.
Nucleic Acids Res., 38, 2010
|
|
3MWK
 
 | | Q28E mutant of HERA N-terminal RecA-like domain, complex with 8-oxo-AMP | | Descriptor: | Heat resistant RNA dependent ATPase, SULFATE ION, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
3NBF
 
 | | Q28E mutant of hera helicase N-terminal domain bound to 8-oxo-ADP | | Descriptor: | Heat resistant RNA dependent ATPase, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl dihydrogen phosphate, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl phosphono hydrogen phosphate | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
1TUT
 
 | | J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns | | Descriptor: | 5'-R(*GP*AP*GP*GP*AP*AP*GP*GP*CP*GP*A)-3', 5'-R(*UP*CP*GP*UP*UP*AP*AP*UP*CP*UP*C)-3' | | Authors: | Znosko, B.M, Kennedy, S.D, Wille, P.C, Krugh, T.R, Turner, D.H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Features and Thermodynamics of the J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns.
Biochemistry, 43, 2004
|
|
2DCU
 
 | | Crystal structure of translation initiation factor aIF2betagamma heterodimer with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Translation initiation factor 2 beta subunit, ... | | Authors: | Sokabe, M, Yao, M, Sakai, N, Toya, S, Tanaka, I. | | Deposit date: | 2006-01-16 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5Y87
 
 | |
1RSN
 
 | | RIBONUCLEASE (RNASE SA) (E.C.3.1.4.8) COMPLEXED WITH EXO-2',3'-CYCLOPHOSPHOROTHIOATE | | Descriptor: | GUANOSINE-2',3'-CYCLOPHOSPHOROTHIOATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Dauter, Z, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of ribonuclease Sa with a cyclic nucleotide and a proposed model for the reaction intermediate.
Eur.J.Biochem., 216, 1993
|
|
5Y85
 
 | |
4CGS
 
 | | Crystal structure of the N-terminal domain of the PA subunit of Dhori virus polymerase | | Descriptor: | GLYCEROL, POLYMERASE SUBUNIT PA | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-11-26 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
7JS8
 
 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH AN ETHYL KETONE INHIBITOR CONTAINING A SPIRO-BICYCLIC GROUP (COMPOUND 22) | | Descriptor: | (1S)-N-{(1S)-7,7-dihydroxy-1-[4-(2-methylquinolin-6-yl)-1H-imidazol-2-yl]nonyl}-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-08-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity.
J.Med.Chem., 64, 2021
|
|
