1MKH
 
 | |
1GU2
 
 | | Crystal structure of oxidized cytochrome c'' from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Enguita, F.J, Pohl, E, Rodrigues, A, Santos, H, Carrondo, M.A. | | Deposit date: | 2002-01-22 | | Release date: | 2003-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
5SFW
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n4c(CC)n1c(nc(n1)CCc2nc(nn2C)N3CCCC3)c(c4)C, micromolar IC50=0.004899 | | Descriptor: | (4R)-5-ethyl-8-methyl-2-{2-[1-methyl-3-(pyrrolidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}[1,2,4]triazolo[1,5-c]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Lerner, C, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
2XER
 
 | |
4RVK
 
 | | CHK1 kinase domain with diazacarbazole compound 8: N-[3-(6-cyano-9H-pyrrolo[2,3-b:5,4-c']dipyridin-3-yl)phenyl]acetamide | | Descriptor: | N-[3-(6-cyano-9H-pyrrolo[2,3-b:5,4-c']dipyridin-3-yl)phenyl]acetamide, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
4K1X
 
 | | Ferredoxin-NADP(H) Reductase mutant with Ala 266 replaced by Tyr (A266Y) and residues 267-272 deleted. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH:ferredoxin reductase, SULFATE ION | | Authors: | Bortolotti, A, Sanchez-Azqueta, A, Maya, C.M, Velazquez-Campoy, A, Hermoso, J.A, Cortez, N. | | Deposit date: | 2013-04-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminal extension of bacterial flavodoxin-reductases: Involvement in the hydride transfer mechanism from the coenzyme.
Biochim.Biophys.Acta, 1837, 2013
|
|
2O7M
 
 | | The C-terminal loop of the homing endonuclease I-CreI is essential for DNA binding and cleavage. Identification of a novel site for specificity engineering in the I-CreI scaffold | | Descriptor: | DNA endonuclease I-CreI | | Authors: | Prieto, J, Redondo, P, Padro, D, Blanco, F.J, Paques, F, Montoya, G. | | Deposit date: | 2006-12-11 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The C-terminal loop of the homing endonuclease I-CreI is essential for site recognition, DNA binding and cleavage
Nucleic Acids Res., 35, 2007
|
|
5G05
 
 | | Cryo-EM structure of combined apo phosphorylated APC | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
5AGS
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct 3-(Aminomethyl)-4-bromo-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 3-(AMINOMETHYL)-4-BROMO-7-ETHOXYBENZO[C][1,2]OXABOROL-1(3H)-OL-MODIFIED ADENOSINE, LEUCYL-TRNA SYNTHETASE, METHIONINE | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
1LMS
 
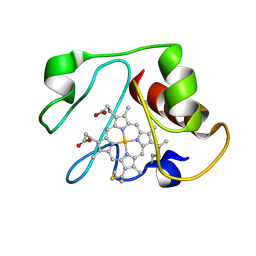 | | Structural model for an alkaline form of ferricytochrome c | | Descriptor: | Cytochrome c, iso-1, HEME C | | Authors: | Assfalg, M, Bertini, I, Dolfi, A, Turano, P, Mauk, A.G, Rosell, F.I, Gray, H.B. | | Deposit date: | 2002-05-02 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural model for an alkaline form of ferricytochrome c
J.Am.Chem.Soc., 125, 2003
|
|
3I5K
 
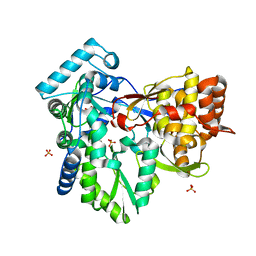 | | Crystal structure of the NS5B polymerase from Hepatitis C Virus (HCV) strain JFH1 | | Descriptor: | PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Simister, P.C, Schmitt, M, Lohmann, V, Bressanelli, S. | | Deposit date: | 2009-07-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of hepatitis C virus strain JFH1 polymerase
J.Virol., 83, 2009
|
|
6IQU
 
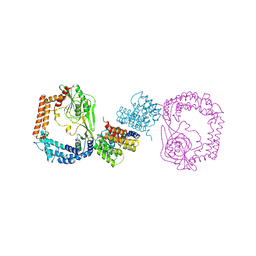 | |
2XLO
 
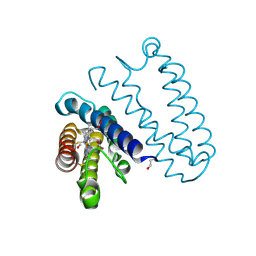 | |
5C2W
 
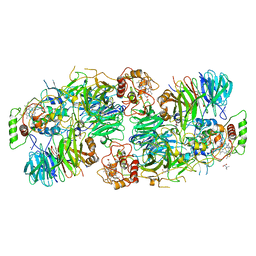 | | Kuenenia stuttgartiensis Hydrazine Synthase Pressurized with 20 bar Xenon | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEME C, ... | | Authors: | Dietl, A, Ferousi, C, Maalcke, W.J, Menzel, A, de Vries, S, Keltjens, J.T, Jetten, M.S.M, Kartal, B, Barends, T.R.M. | | Deposit date: | 2015-06-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The inner workings of the hydrazine synthase multiprotein complex.
Nature, 527, 2015
|
|
2XLV
 
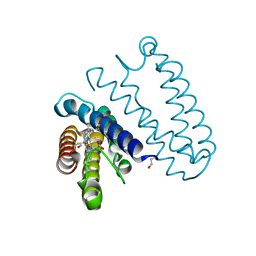 | | Cytochrome c prime from Alcaligenes xylosoxidans: Ferrous R124F variant with bound NO | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R, Hasnain, S.S. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Distal-to-Proximal No Conversion in Hemoproteins: The Role of the Proximal Pocket.
J.Mol.Biol., 405, 2011
|
|
4NUT
 
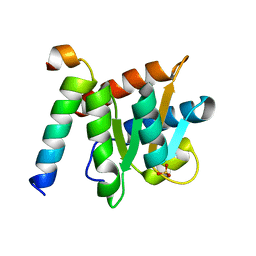 | | Crystal structure of the complex between Snu13p and the PEP domain of Rsa1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, Ribosome assembly 1 protein, SULFATE ION | | Authors: | Charron, C, Chagot, M.E, Manival, X, Branlant, C, Charpentier, B. | | Deposit date: | 2013-12-04 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control
J.Cell Biol., 207, 2014
|
|
2IPQ
 
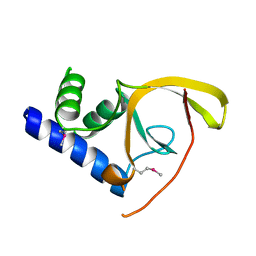 | | Crystal structure of C-terminal domain of Salmonella Enterica protein STY4665, PFAM DUF1528 | | Descriptor: | Hypothetical protein STY4665 | | Authors: | Ramagopal, U.A, Bonanno, J.B, Gilmore, J, Toro, R, Bain, K.T, Reyes, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of C-terminal domain of Hypothetical protein STY4665
To be Published
|
|
4RNJ
 
 | | PaMorA phosphodiesterase domain, apo form | | Descriptor: | Motility regulator | | Authors: | Phippen, C.W, Tews, I. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator.
Febs Lett., 588, 2014
|
|
2JSW
 
 | | Solution Structure of the R13 Domain of Talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Gingras, A.R, Bate, N, Critchley, D.R.C, Barsukov, I.L. | | Deposit date: | 2007-07-17 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the C-terminal actin-binding domain of talin.
Embo J., 27, 2007
|
|
2JVL
 
 | | NMR structure of the C-terminal domain of MBF1 of Trichoderma reesei | | Descriptor: | TrMBF1 | | Authors: | Kopke Salinas, R, Tomaselli, S, Camilo, C.M, Valencia, E.Y, Farah, C.S, El-Dorry, H, Chambergo, F.S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of multiprotein bridging factor 1 (MBF1) of Trichoderma reesei.
Proteins, 75, 2009
|
|
5AGT
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-4-chloro-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, GLYCEROL, LEUCINE--TRNA LIGASE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
7BBT
 
 | | Structure of cytochrome c in complex with a p-benzyl-sulfonato-calix[8]arene-PEG pseudorotaxane | | Descriptor: | 4-[[49,50,51,52,53,54,55,56-octahydroxy-11,17,23,29,35,41,47-heptakis[(4-sulfonatophenyl)methyl]-5-nonacyclo[43.3.1.13,7.19,13.115,19.121,25.127,31.133,37.139,43]hexapentaconta-1(49),3,5,7(56),9,11,13(55),15,17,19(54),21,23,25(53),27,29,31(52),33,35,37(51),39,41,43(50),45,47-tetracosaenyl]methyl]benzenesulfonate, Cytochrome c iso-1, HEME C, ... | | Authors: | Mockler, N.M, Ramberg, K, Guagnini, F, Raston, C.L, Crowley, P.B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Noncovalent Protein-Pseudorotaxane Assembly Incorporating an Extended Arm Calix[8]arene with alpha-Helical Recognition Properties.
Cryst.Growth Des., 21, 2021
|
|
3IA8
 
 | |
5G04
 
 | | Structure of the human APC-Cdc20-Hsl1 complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
5C2V
 
 | | Kuenenia stuttgartiensis Hydrazine Synthase | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEME C, ... | | Authors: | Dietl, A, Ferousi, C, Maalcke, W.J, Menzel, A, de Vries, S, Keltjens, J.T, Jetten, M.S.M, Kartal, B, Barends, T.R.M. | | Deposit date: | 2015-06-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The inner workings of the hydrazine synthase multiprotein complex.
Nature, 527, 2015
|
|
