1ZEQ
 
 | | 1.5 A Structure of apo-CusF residues 6-88 from Escherichia coli | | Descriptor: | Cation efflux system protein cusF | | Authors: | Loftin, I.R, Franke, S, Roberts, S.A, Weichsel, A, Heroux, A, Montfort, W.R, Rensing, C, McEvoy, M.M. | | Deposit date: | 2005-04-19 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Novel Copper-Binding Fold for the Periplasmic Copper Resistance Protein CusF.
Biochemistry, 44, 2005
|
|
1UY4
 
 | |
4O0A
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-{4-[({[4-(5-carboxyfuran-2-yl)-2-chlorophenyl]carbonothioyl}amino)methyl]phenyl}-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejniczak, E.T, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-12-13 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J. Med. Chem., 56, 2013
|
|
1UY3
 
 | |
3FKU
 
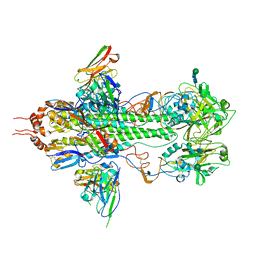 | | Crystal structure of influenza hemagglutinin (H5) in complex with a broadly neutralizing antibody F10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Hwang, W.C, Santelli, E, Stec, B, Wei, G, Cadwell, G, Bankston, L.A, Sui, J, Perez, S, Aird, D, Chen, L.M, Ali, M, Murakami, A, Yammanuru, A, Han, T, Cox, N, Donis, R.O, Liddington, R.C, Marasco, W.A. | | Deposit date: | 2008-12-17 | | Release date: | 2009-02-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1VC5
 
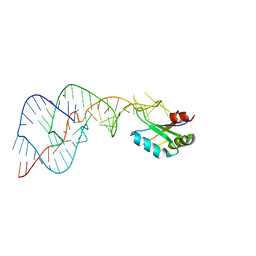 | | Crystal Structure of the Wild Type Hepatitis Delta Virus Gemonic Ribozyme Precursor, in EDTA solution | | Descriptor: | Hepatitis Delta virus ribozyme, SODIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1X2H
 
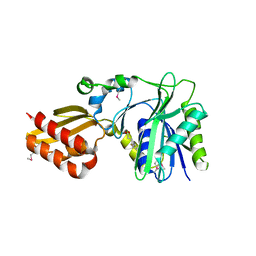 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli complexed with lipoic acid | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1X2G
 
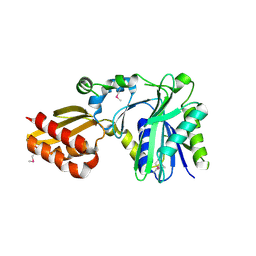 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli | | Descriptor: | Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
2EWJ
 
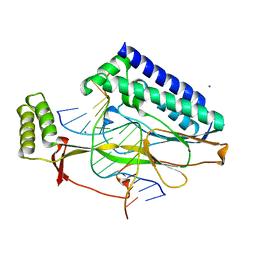 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With DNA- Locked form | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*G*AP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2005-11-03 | | Release date: | 2006-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular mousetrap determines polarity of termination of DNA replication in E. coli.
Cell(Cambridge,Mass.), 125, 2006
|
|
4FBX
 
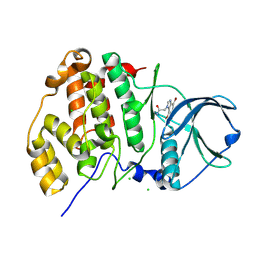 | | Complex structure of human protein kinase CK2 catalytic subunit crystallized in the presence of a bisubstrate inhibitor | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha, bisubstrate inhibitor | | Authors: | Enkvist, E, Viht, K, Bischoff, N, Vahter, J, Saaver, S, Raidaru, G, Issinger, O.-G, Niefind, K, Uri, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A subnanomolar fluorescent probe for protein kinase CK2 interaction studies.
Org.Biomol.Chem., 10, 2012
|
|
1THB
 
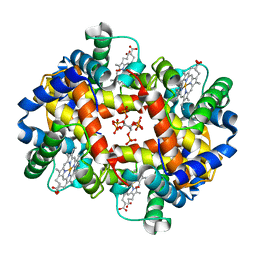 | |
4RQY
 
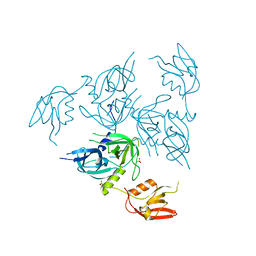 | |
2XNE
 
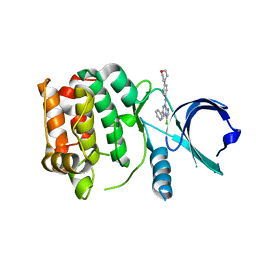 | | Structure of Aurora-A bound to an imidazopyrazine inhibitor | | Descriptor: | 3-chloro-N-(4-morpholin-4-ylphenyl)-6-pyridin-3-ylimidazo[1,2-a]pyrazin-8-amine, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
3NE8
 
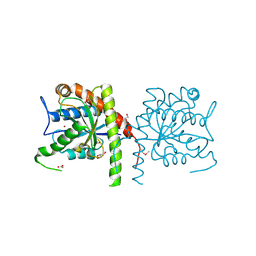 | | The crystal structure of a domain from N-acetylmuramoyl-l-alanine amidase of Bartonella henselae str. Houston-1 | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Rakowski, E, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division.
Mol.Microbiol., 85, 2012
|
|
2B4B
 
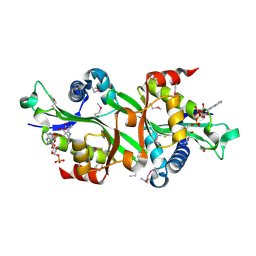 | | SSAT+COA+BE-3-3-3, K26R mutant | | Descriptor: | COENZYME A, Diamine acetyltransferase 1, N-ETHYL-N-[3-(PROPYLAMINO)PROPYL]PROPANE-1,3-DIAMINE | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-23 | | Release date: | 2006-01-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2XNG
 
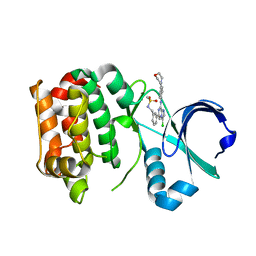 | | Structure of Aurora-A bound to a selective imidazopyrazine inhibitor | | Descriptor: | N-(3-{3-chloro-8-[(4-morpholin-4-ylphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzyl)methanesulfonamide, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
2XCQ
 
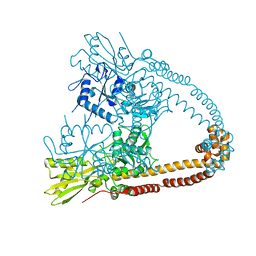 | | The 2.98A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
4LUZ
 
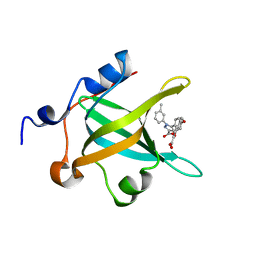 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]oxy}phenyl)-1-(3-methylphenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
2A06
 
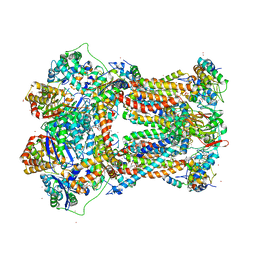 | | Bovine cytochrome bc1 complex with stigmatellin bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, CARDIOLIPIN, ... | | Authors: | Huang, L.S, Cobessi, D, Tung, E.Y, Berry, E.A. | | Deposit date: | 2005-06-16 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the Respiratory Chain Inhibitor Antimycin to the Mitochondrial bc(1) Complex: A New Crystal Structure Reveals an Altered Intramolecular Hydrogen-bonding Pattern.
J.Mol.Biol., 351, 2005
|
|
1JX2
 
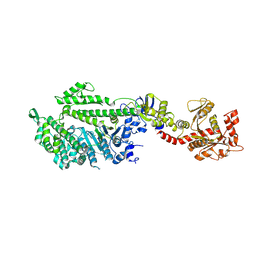 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE-FREE DYNAMIN A GTPASE DOMAIN, DETERMINED AS MYOSIN FUSION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin-2 heavy chain,Dynamin-A, ... | | Authors: | Niemann, H.H, Knetsch, M.L.W, Scherer, A, Manstein, D.J, Kull, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2001-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms.
EMBO J., 20, 2001
|
|
1RZT
 
 | | Crystal structure of DNA polymerase lambda complexed with a two nucleotide gap DNA molecule | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*GP*GP*CP*AP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*G)-3', ... | | Authors: | Pedersen, L.C, Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Blanco, L, Kunkel, T.A. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology.
Mol.Cell, 13, 2004
|
|
3K6M
 
 | | Dynamic domains of Succinyl-CoA:3-ketoacid-coenzyme A transferase from pig heart. | | Descriptor: | CHLORIDE ION, GLYCEROL, Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, ... | | Authors: | Coker, S, Lloyd, A, Mitchell, E, Lewis, G.R, Shoolingin-Jordan, P, Coker, A.R. | | Deposit date: | 2009-10-09 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The high-resolution structure of pig heart succinyl-CoA:3-oxoacid coenzyme A transferase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4G2M
 
 | |
2AAV
 
 | | Solution NMR structure of Filamin A domain 17 | | Descriptor: | Filamin A | | Authors: | Nakamura, F, Pudas, R, Heikkinen, O, Permi, P, Kilpelainen, I, Munday, A.D, Hartwig, J.H, Stossel, T.P, Ylanne, J. | | Deposit date: | 2005-07-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the GPIb-filamin A complex
Blood, 107, 2006
|
|
4U28
 
 | | Crystal structure of apo Phosphoribosyl isomerase A from Streptomyces sviceus ATCC 29083 | | Descriptor: | PHOSPHATE ION, Phosphoribosyl isomerase A | | Authors: | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
