7ZVD
 
 | | K89 acetylated glucose-6-phosphate dehydrogenase (G6PD) in a complex with structural NADP+ | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wu, F, Muskat, N.H, Nudelman, H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
7ZTN
 
 | | Crystal structure of fungal CE16 acetyl xylan esterase | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Kosinas, C, Pentari, C, Zerva, A, Topakas, E. | | Deposit date: | 2022-05-10 | | Release date: | 2023-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The role of CE16 exo-deacetylases in hemicellulolytic enzyme mixtures revealed by the biochemical and structural study of the novel TtCE16B esterase.
Carbohydr Polym, 327, 2024
|
|
5LJX
 
 | |
8A0S
 
 | | MTH1 in complex with TH013350 | | Descriptor: | 4-[(2-chlorophenyl)methylsulfanyl]-5~{H}-pyrimidin-2-one, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-05-30 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | MTH1 in complex with TH013350
To Be Published
|
|
6U0S
 
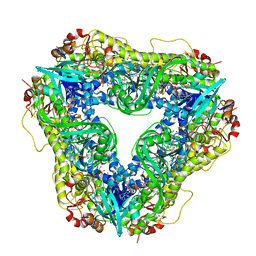 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | Authors: | Shi, R, Manenda, M. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6ASL
 
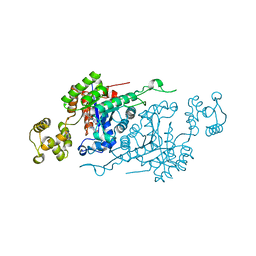 | |
6U1M
 
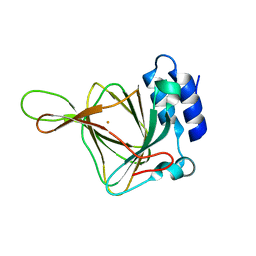 | |
2ZYS
 
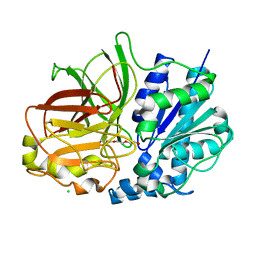 | | A. Fulgidus lipase with fatty acid fragment and chloride | | Descriptor: | CHLORIDE ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
8PQN
 
 | | NQO1 bound to RBS-10 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1, ~{N}-[4-[(3-methylphenyl)carbonylamino]phenyl]-5-nitro-furan-2-carboxamide | | Authors: | Pous, J, Jose-Duran, F, Mayor-Ruiz, C, Riera, A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Discovery and Mechanistic Elucidation of NQO1-Bioactivatable Small Molecules That Overcome Resistance to Degraders.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PTE
 
 | | Polyoxidovanadate interaction with proteins: crystal structure of lysozyme bound to octadecavanadate ion (structure B) | | Descriptor: | Lysozyme C, NITRATE ION, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Ferraro, G, Merlino, A. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Stabilization and Binding of [V 4 O 12 ] 4- and Unprecedented [V 20 O 54 (NO 3 )] n- to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug V IV O(acetylacetonato) 2.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8A3A
 
 | | MTH1 in complex with TH013074 | | Descriptor: | 4-(4-methylphenyl)-1~{H}-quinazolin-2-one, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-06-07 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 in complex with TH013074
To Be Published
|
|
1F2F
 
 | | SRC SH2 THREF1TRP MUTANT | | Descriptor: | PHOSPHATE ION, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Kimber, M.S, Nachman, J, Cunningham, A.M, Gish, G.D, Pawson, T, Pai, E.F. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for specificity switching of the Src SH2 domain.
Mol.Cell, 5, 2000
|
|
8D7F
 
 | | FlvF from Aspergillus flavus in complex with Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Terpene cyclase flvF | | Authors: | Tararina, M.A, Christianson, D.W. | | Deposit date: | 2022-06-07 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the Repurposed Fungal Terpene Cyclase FlvF Implicated in the C-N Bond-Forming Reaction of Flavunoidine Biosynthesis.
Biochemistry, 61, 2022
|
|
8A1U
 
 | | Sodium pumping NADH-quinone oxidoreductase with substrates NADH and Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6KZT
 
 | | Crystal structure of cytochrome P450mel 107F1 with biaryl coupling reactivity | | Descriptor: | Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hou, X.D, Rao, Y.J, Icyishaka, P, Li, C.X, Lou, J.L. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and X-ray analysis of cytochrome P450mel 107F1 with biaryl coupling reactivity
To Be Published
|
|
6U4J
 
 | | Crystal structure of IDH1 R132H mutant in complex with FT-2102 | | Descriptor: | 5-{[(1S)-1-(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)ethyl]amino}-1-methyl-6-oxo-1,6-dihydropyridine-2-carbonitrile, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-08-25 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Based Design and Identification of FT-2102 (Olutasidenib), a Potent Mutant-Selective IDH1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
8ILJ
 
 | |
8A34
 
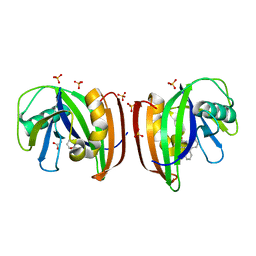 | | MTH1 in complex with TH013071 | | Descriptor: | 5-(2-phenylphenyl)-1H-pyrimidine-2,4-dione, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-06-07 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MTH1 in complex with TH013071
To Be Published
|
|
6AUK
 
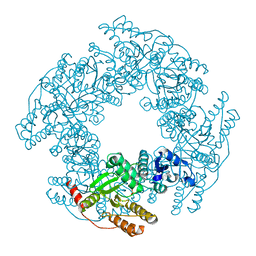 | |
1F35
 
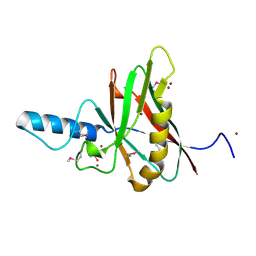 | |
8A07
 
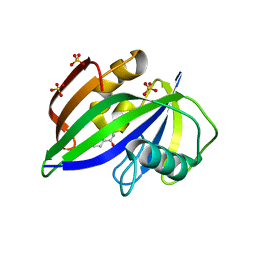 | |
8A0T
 
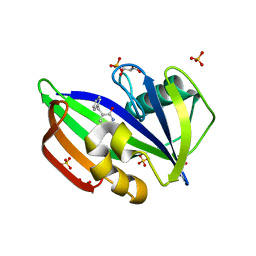 | | MTH1 in complex with TH012532 | | Descriptor: | 5-(phenylmethyl)pyrimidine-2,4-diol, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Scaletti, E, Stenmark, P. | | Deposit date: | 2022-05-30 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MTH1 in complex with TH012532
To Be Published
|
|
8PF8
 
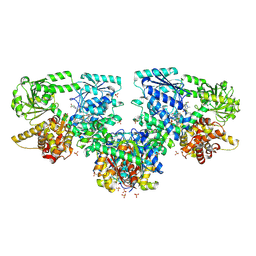 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-72 | | Descriptor: | (2~{R})-3-bis[2-methyl-5-(trifluoromethyl)pyrazol-3-yl]boranyloxypropane-1,2-diol, GLYCEROL, Probable fatty oxidation protein FadB, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
5LN5
 
 | |
3AB0
 
 | |
