5LWI
 
 | | Israeli acute paralysis virus heated to 63 degree - empty particle | | Descriptor: | Structural polyprotein, VP1, VP2 | | Authors: | Mullapudi, E, Fuzik, T, Pridal, A, Plevka, P. | | Deposit date: | 2016-09-16 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-electron Microscopy Study of the Genome Release of the Dicistrovirus Israeli Acute Bee Paralysis Virus.
J. Virol., 91, 2017
|
|
5LH6
 
 | | High dose Thaumatin - 360-400 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LM7
 
 | | Crystal structure of the lambda N-Nus factor complex | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | Authors: | Said, N, Santos, K, Weber, G, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
5LMS
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2C) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LP4
 
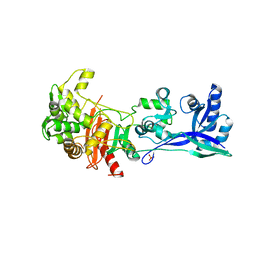 | | Penicillin-Binding Protein (PBP2) from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), SULFATE ION | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
5LH1
 
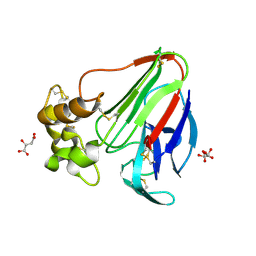 | | Low dose Thaumatin - 360-400 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LHH
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-ethyl-N-[3-(methoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-ethyl-~{N}-[3-(methoxymethyl)-2-[[4-[[(3~{R})-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
5LUE
 
 | | Minor form of the recombinant cytotoxin-1 from N. oxiana | | Descriptor: | VC-1=CYTOTOXIN | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
5LUT
 
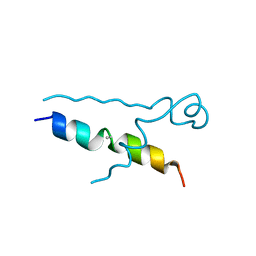 | | Structures of DHBN domain of Gallus gallus BLM helicase | | Descriptor: | BLM helicase, PHOSPHATE ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5FB6
 
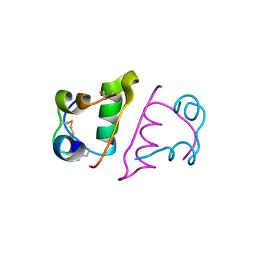 | | Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering | | Descriptor: | Insulin Chain A, Insulin Chain B | | Authors: | Roedig, P, Duman, R, Sanchez-Weatherby, J, Vartiainen, I, Burkhardt, A, Warmer, M, David, C, Wagner, A, Meents, A. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering.
J.Appl.Crystallogr., 49, 2016
|
|
5LY6
 
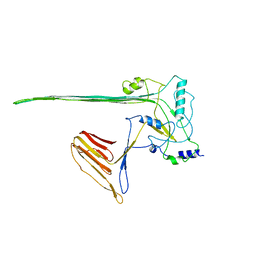 | | CryoEM structure of the membrane pore complex of Pneumolysin at 4.5A | | Descriptor: | Pneumolysin | | Authors: | van Pee, K, Neuhaus, A, D'Imprima, E, Mills, D.J, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2016-09-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin.
Elife, 6, 2017
|
|
5LYG
 
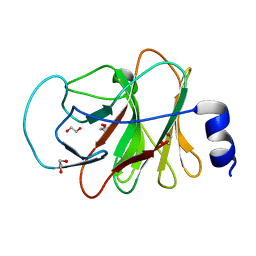 | | CRYSTAL STRUCTURE OF INTRACELLULAR B30.2 DOMAIN OF BTN3A1 BOUND TO MALONATE | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, MALONATE ION | | Authors: | Mohammed, F, Baker, A.T, Salim, M, Willcox, B.E. | | Deposit date: | 2016-09-28 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | BTN3A1 Discriminates gamma delta T Cell Phosphoantigens from Nonantigenic Small Molecules via a Conformational Sensor in Its B30.2 Domain.
ACS Chem. Biol., 12, 2017
|
|
5LN3
 
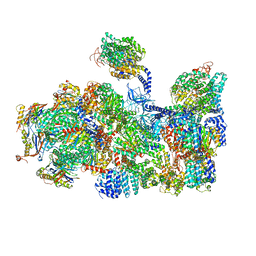 | | The human 26S Proteasome at 6.8 Ang. | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | Deposit date: | 2016-08-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
7VOJ
 
 | | Al-bound structure of the AtALMT1 mutant M60A | | Descriptor: | ACETIC ACID, ALUMINUM ION, Aluminum-activated malate transporter 1 | | Authors: | Wang, J. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis.
Cell Res., 32, 2022
|
|
5LUP
 
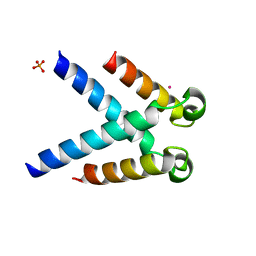 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | BLM protein, PHOSPHATE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
7VQ5
 
 | |
5LKH
 
 | | Cryo-EM structure of the Tc toxin TcdA1 in its pore state (obtained by flexible fitting) | | Descriptor: | TcdA1 | | Authors: | Gatsogiannis, C, Merino, F, Prumbaum, D, Roderer, D, Leidreiter, F, Meusch, D, Raunser, S. | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Membrane insertion of a Tc toxin in near-atomic detail.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5LMR
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2B) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5M56
 
 | | Monoclinic complex structure of human protein kinase CK2 catalytic subunit (isoform CK2alpha') with the inhibitor 4'-carboxy-6,8-chloro-flavonol (FLC21) | | Descriptor: | 4-[6,8-bis(chloranyl)-3-oxidanyl-4-oxidanylidene-chromen-2-yl]benzoic acid, CHLORIDE ION, Casein kinase II subunit alpha', ... | | Authors: | Niefind, K, Bischoff, N, Yarmoluk, S.M, Bdzhola, V.G, Golub, A.G, Balanda, A.O, Prykhod'ko, A.O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.237 Å) | | Cite: | Structural Hypervariability of the Two Human Protein Kinase CK2 Catalytic Subunit Paralogs Revealed by Complex Structures with a Flavonol- and a Thieno[2,3-d]pyrimidine-Based Inhibitor.
Pharmaceuticals, 10, 2017
|
|
5LM9
 
 | | Structure of E. coli NusA | | Descriptor: | MAGNESIUM ION, SULFATE ION, Transcription termination/antitermination protein NusA | | Authors: | Said, N, Weber, G, Santos, K, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
7V3H
 
 | | DENV2_NGC_Fab_C10 28degrees (3Fab:3E) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C10 IgG heavy chain variable region, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
7V3I
 
 | | DENV2_NGC_Fab_C10 4degrees (3Fab:3E) | | Descriptor: | Envelope protein E, Fab_C10_heavy_chain, Fab_C10_light_chain, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
7VSG
 
 | |
7V3J
 
 | | DENV2:F(ab')2-local | | Descriptor: | Envelope protein E, Fab_C10_heavy_chain, Fab_C10_light_chain, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
7VSH
 
 | |
