4DG8
 
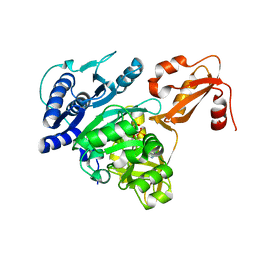 | | Structure of PA1221, an NRPS protein containing adenylation and PCP domains | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE MONOPHOSPHATE, PA1221 | | Authors: | Mitchell, C.A, Shi, C, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of PA1221, a Nonribosomal Peptide Synthetase Containing Adenylation and Peptidyl Carrier Protein Domains.
Biochemistry, 51, 2012
|
|
4DN3
 
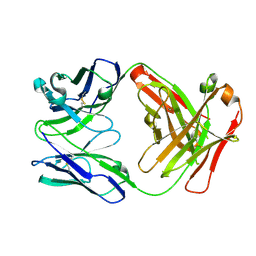 | | Crystal structure of anti-mcp-1 antibody cnto888 | | Descriptor: | CNTO888 HEAVY CHAIN, CNTO888 LIGHT CHAIN | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
7A0Z
 
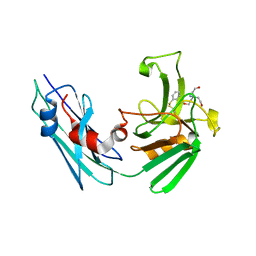 | | LppS with covalent adduct derived from 1b | | Descriptor: | L,D-transpeptidase 2, TRIS(HYDROXYETHYL)AMINOMETHANE, benzenethiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7P4U
 
 | |
7P85
 
 | |
3SP6
 
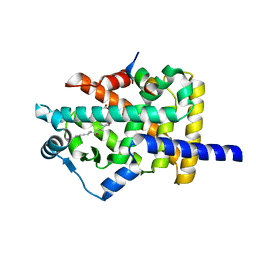 | | Structural basis for iloprost as a dual PPARalpha/delta agonist | | Descriptor: | (5E)-5-[(3aS,4R,5R,6aS)-5-hydroxy-4-[(1E,3S,4R)-3-hydroxy-4-methyloct-1-en-6-yn-1-yl]hexahydropentalen-2(1H)-ylidene]pentanoic acid, Peroxisome proliferator-activated receptor alpha, Peroxisome proliferator-activated receptor gamma coactivator 1-beta | | Authors: | Rong, H, Li, Y. | | Deposit date: | 2011-07-01 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for iloprost as a dual peroxisome proliferator-activated receptor alpha/delta agonist.
J.Biol.Chem., 286, 2011
|
|
1FSR
 
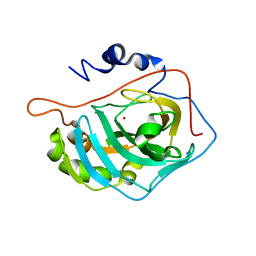 | | X-RAY CRYSTAL STRUCTURE OF COPPER-BOUND F93S/F95L/W97M CARBONIC ANHYDRASE (CAII) VARIANT | | Descriptor: | CARBONIC ANHYDRASE II, COPPER (II) ION | | Authors: | Cox, J.D, Hunt, J.A, Compher, K.M, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2000-09-11 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II.
Biochemistry, 39, 2000
|
|
1FQL
 
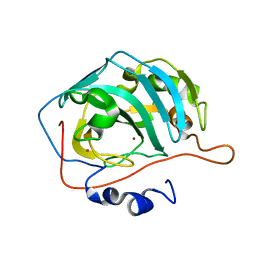 | | X-RAY CRYSTAL STRUCTURE OF ZINC-BOUND F95M/W97V CARBONIC ANHYDRASE (CAII) VARIANT | | Descriptor: | CARBONIC ANHYDRASE, MERCURY (II) ION, ZINC ION | | Authors: | Cox, J.D, Hunt, J.A, Compher, K.M, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2000-09-05 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II.
Biochemistry, 39, 2000
|
|
4JSC
 
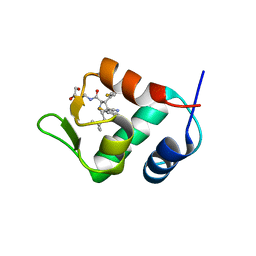 | | The 2.5A crystal structure of humanized Xenopus MDM2 with RO5316533 - a pyrrolidine MDM2 inhibitor | | Descriptor: | (3S,4R,5S)-3-(3-chloro-2-fluorophenyl)-4-(4-chloro-2-fluorophenyl)-4-cyano-N-[(3S)-3,4-dihydroxybutyl]-5-(2,2-dimethylpropyl)-D-prolinamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Janson, C.A, Lukacs, C, Graves, B. | | Deposit date: | 2013-03-22 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of RG7388, a Potent and Selective p53-MDM2 Inhibitor in Clinical Development.
J.Med.Chem., 56, 2013
|
|
2AQD
 
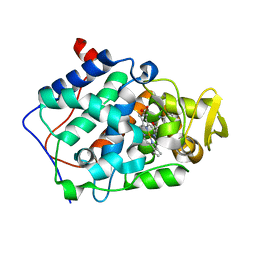 | | cytochrome c peroxidase (CCP) in complex with 2,5-diaminopyridine | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brenk, R, Vetter, S.W, Boyce, S.E, Goodin, D.B, Shoichet, B.K. | | Deposit date: | 2005-08-17 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Probing molecular docking in a charged model binding site.
J.Mol.Biol., 357, 2006
|
|
3JZF
 
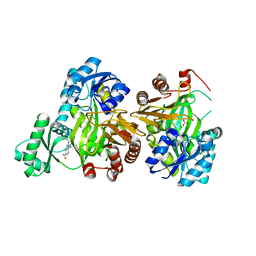 | |
6UL3
 
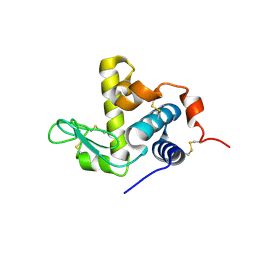 | |
7BC2
 
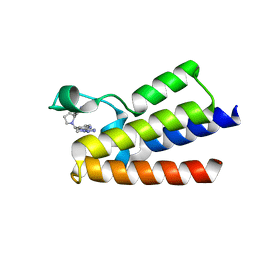 | | BAZ2A bromodomain in complex with triazole compound MS04-TN04 | | Descriptor: | 2-(1-methyl-1H-1,2,3-triazol-5-yl)-5-((2-(pyridin-4-yl)pyrrolidin-1-yl)methyl)-1H-benzo[d]imidazole, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
6TO3
 
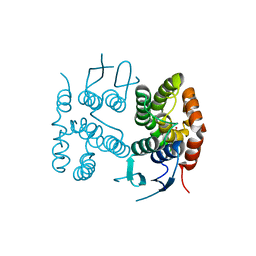 | |
3QP5
 
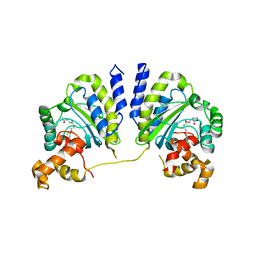 | | Crystal structure of CviR bound to antagonist chlorolactone (CL) | | Descriptor: | 4-(4-chlorophenoxy)-N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, CviR transcriptional regulator | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.249 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
1FQR
 
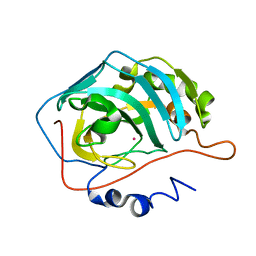 | | X-RAY CRYSTAL STRUCTURE OF COBALT-BOUND F93I/F95M/W97V CARBONIC ANHYDRASE (CAII) VARIANT | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Cox, J.D, Hunt, J.A, Compher, K.M, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2000-09-06 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II.
Biochemistry, 39, 2000
|
|
1D8G
 
 | | ULTRAHIGH RESOLUTION CRYSTAL STRUCTURE OF B-DNA DECAMER D(CCAGTACTGG) | | Descriptor: | 5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*GP*)-3', CALCIUM ION | | Authors: | Kielkopf, C.L, Ding, S, Kuhn, P, Rees, D.C. | | Deposit date: | 1999-10-23 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Conformational flexibility of B-DNA at 0.74 A resolution: d(CCAGTACTGG)(2).
J.Mol.Biol., 296, 2000
|
|
1WN3
 
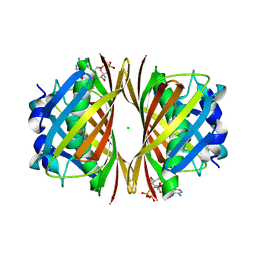 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, CHLORIDE ION, HEXANOYL-COENZYME A, ... | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
8U3Z
 
 | | Model of TTLL6 bound to microtubule from composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 2024
|
|
4FIT
 
 | | FHIT-APO | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
6S2Z
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Brassica oleracea var. Botrytis with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-Soluble Chlorophyll Protein | | Authors: | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | Deposit date: | 2019-06-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
6UKC
 
 | |
8X17
 
 | | Cryo-EM structure of adenosine receptor A3AR bound to CF102 | | Descriptor: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of adenosine receptor A 3 AR bound to selective agonists.
Nat Commun, 15, 2024
|
|
8X16
 
 | | Cryo-EM structure of adenosine receptor A3AR bound to CF101 | | Descriptor: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures of adenosine receptor A 3 AR bound to selective agonists.
Nat Commun, 15, 2024
|
|
1FWU
 
 | |
