2BJH
 
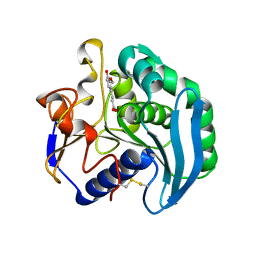 | | Crystal Structure Of S133A AnFaeA-ferulic acid complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Faulds, C.B, Molina, R, Gonzalez, R, Husband, F, Juge, N, Sanz-Aparicio, J, Hermoso, J.A. | | Deposit date: | 2005-02-02 | | Release date: | 2005-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Probing the Determinants of Substrate Specificity of a Feruloyl Esterase, Anfaea, from Aspergillus Niger
FEBS J., 272, 2005
|
|
3MTN
 
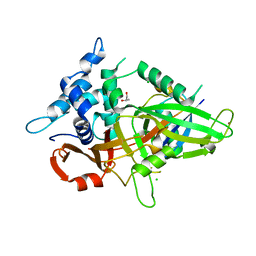 | | Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, UBIQUITIN VARIANT UBV.21.4, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
1EIS
 
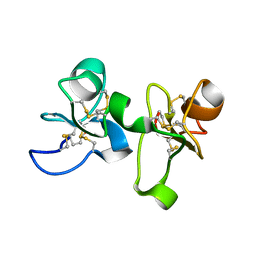 | | UDA UNCOMPLEXED FORM. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | PROTEIN (AGGLUTININ ISOLECTIN VI/AGGLUTININ ISOLECTIN V) | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-02-28 | | Release date: | 2000-06-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1ENM
 
 | | UDA TRISACCHARIDE COMPLEX. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN I/AGGLUTININ ISOLECTIN V/ AGGLUTININ ISOLECTIN VI | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
3IRR
 
 | | Crystal Structure of a Z-Z junction (with HEPES intercalating) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*A*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*G*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6ARB
 
 | | CRYSTAL STRUCTURE OF PROTEIN CitE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, PYRUVATE AND COENZYME A | | Descriptor: | COENZYME A, Citrate lyase subunit beta-like protein, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3B8S
 
 | | Crystal structure of wild-type chitinase A from Vibrio harveyi | | Descriptor: | Chitinase A | | Authors: | Songsiriritthigul, C, Pantoom, S, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
5I4L
 
 | | Crystal structure of Amicoumacin A bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Prokhorova, I.V, Yusupova, G, Yusupov, M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Amicoumacin A induces cancer cell death by targeting the eukaryotic ribosome.
Sci Rep, 6, 2016
|
|
1UY3
 
 | |
5TBO
 
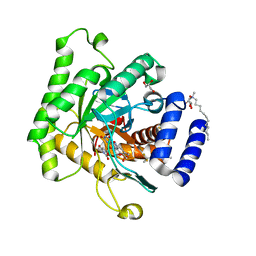 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM421 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[6-(trifluoromethyl)pyridin-3-yl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M. | | Deposit date: | 2016-09-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | A Triazolopyrimidine-Based Dihydroorotate Dehydrogenase Inhibitor with Improved Drug-like Properties for Treatment and Prevention of Malaria.
ACS Infect Dis, 2, 2016
|
|
6I52
 
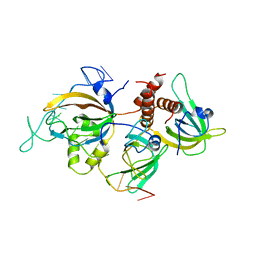 | | Yeast RPA bound to ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Replication factor A protein 1, Replication factor A protein 2, ... | | Authors: | Yates, L.A, Aramayo, R.J, Zhang, X. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA.
Nat Commun, 9, 2018
|
|
4KLQ
 
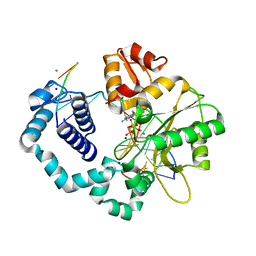 | | Observing a DNA polymerase choose right from wrong. | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*A)-3', ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
3BS1
 
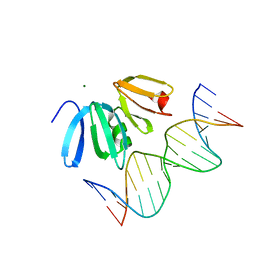 | | Structure of the Staphylococcus aureus AgrA LytTR Domain Bound to DNA Reveals a Beta Fold with a Novel Mode of Binding | | Descriptor: | Accessory gene regulator protein A, DNA (5'-D(*DAP*DAP*(BRU)P*DAP*DCP*DTP*DTP*DAP*DAP*DCP*DTP*DGP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DTP*DAP*DAP*DCP*DAP*DGP*DTP*DTP*DAP*DAP*DGP*(BRU)P*DAP*DT)-3'), ... | | Authors: | Sidote, D.J, Barbieri, C, Wu, T, Stock, A.M. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Staphylococcus aureus AgrA LytTR Domain Bound to DNA Reveals a Beta Fold with an Unusual Mode of Binding.
Structure, 16, 2008
|
|
1UY1
 
 | |
1B5F
 
 | | NATIVE CARDOSIN A FROM CYNARA CARDUNCULUS L. | | Descriptor: | PROTEIN (CARDOSIN A), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frazao, C, Bento, I, Carrondo, M.A. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp-containing aspartic proteinase from the flowers of Cynara cardunculus L.
J.Biol.Chem., 274, 1999
|
|
1OE9
 
 | | Crystal structure of Myosin V motor with essential light chain-nucleotide-free | | Descriptor: | MYOSIN LIGHT CHAIN 1, SLOW-TWITCH MUSCLE A ISOFORM, MYOSIN VA, ... | | Authors: | Coureux, P.-D, Wells, A.L, Menetrey, J, Yengo, C.M, Morris, C.A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2003-03-21 | | Release date: | 2003-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural State of the Myosin V Motor without Bound Nucleotide
Nature, 425, 2003
|
|
3MGT
 
 | | Crystal structure of a H5-specific CTL epitope variant derived from H5N1 influenza virus in complex with HLA-A*0201 | | Descriptor: | 10-meric peptide from Hemagglutinin, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Sun, Y, Liu, J, Yang, M, Gao, F, Zhou, J, Kitamura, Y. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses
J.Gen.Virol., 91, 2010
|
|
1YY7
 
 | | Crystal structure of stringent starvation protein A (SspA), an RNA polymerase-associated transcription factor | | Descriptor: | CITRIC ACID, stringent starvation protein A | | Authors: | Hansen, A.-M, Gu, Y, Li, M, Andrykovitch, M, Waugh, D.S, Jin, D.J, Ji, X. | | Deposit date: | 2005-02-23 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the function of stringent starvation protein A as a transcription factor
J.Biol.Chem., 280, 2005
|
|
4C93
 
 | |
3B9D
 
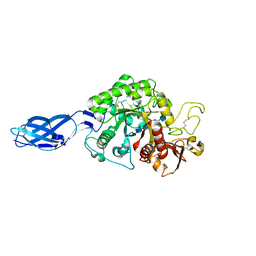 | | Crystal structure of Vibrio harveyi chitinase A complexed with pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Songsiriritthigul, C, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
4PKD
 
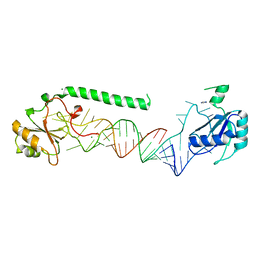 | | U1-70k in complex with U1 snRNA stem-loops 1 and U1-A RRM in complex with stem-loop 2 | | Descriptor: | IMIDAZOLE, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A,U1 small nuclear ribonucleoprotein 70 kDa, ... | | Authors: | Oubridge, C, Kondo, Y, van Roon, A.M, Nagai, K. | | Deposit date: | 2014-05-14 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition.
Elife, 4, 2015
|
|
3B9E
 
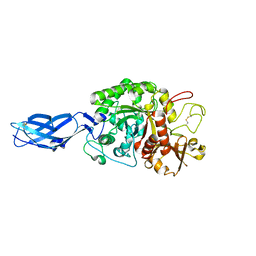 | | Crystal structure of inactive mutant E315M chitinase A from Vibrio harveyi | | Descriptor: | Chitinase A | | Authors: | Songsiriritthigul, C, Pantoom, S, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
3V00
 
 | |
3H0E
 
 | | 3,4-Dihydropyrimido(1,2-a)indol-10(2H)-ones as Potent Non-Peptidic Inhibitors of Caspase-3 | | Descriptor: | (10S)-3,3-dimethyl-8-{[(2S)-2-(phenoxymethyl)pyrrolidin-1-yl]sulfonyl}-2,3,4,10-tetrahydropyrimido[1,2-a]indol-10-ol, Caspase-3 | | Authors: | Xu, W. | | Deposit date: | 2009-04-09 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | 3,4-Dihydropyrimido(1,2-a)indol-10(2H)-ones as potent non-peptidic inhibitors of caspase-3
Bioorg.Med.Chem., 17, 2009
|
|
6AS5
 
 | | CRYSTAL STRUCTURE OF PROTEIN CitE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, ACETOACETATE AND COENZYME A | | Descriptor: | ACETOACETIC ACID, COENZYME A, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
