5FH4
 
 | |
5FKY
 
 | | Structure of a hydrolase bound with an inhibitor | | Descriptor: | (3aR,5R,6S,7R,7aR)-2-amino-5-(hydroxymethyl)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, GLYCEROL, O-GLCNACASE BT_4395 | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
8CT5
 
 | |
1KHB
 
 | | PEPCK complex with nonhydrolyzable GTP analog, native data | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
3TPK
 
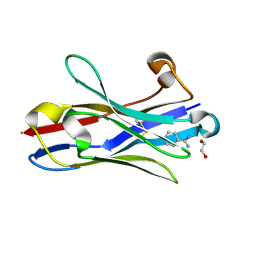 | | Crystal structure of the oligomer-specific KW1 antibody fragment | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, Immunoglobulin heavy chain antibody variable domain KW1 | | Authors: | Parthier, C, Morgado, I, Stubbs, M.T, Fandrich, M. | | Deposit date: | 2011-09-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of beta-amyloid oligomer recognition with a conformational antibody fragment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8C8Q
 
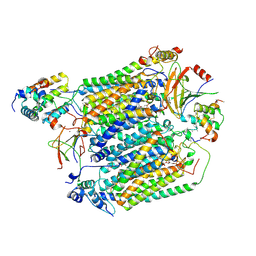 | | Cytochrome c oxidase from Schizosaccharomyces pombe | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cytochrome c oxidase polypeptide 5, ... | | Authors: | Moe, A, Adelroth, P, Brzezinski, P, Nasvik Ojemyr, L. | | Deposit date: | 2023-01-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structure and function of S. pombe complex IV with bound respiratory supercomplex factor.
Commun Chem, 6, 2023
|
|
5FTW
 
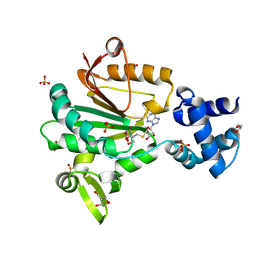 | | Crystal structure of glutamate O-methyltransferase in complex with S- adenosyl-L-homocysteine (SAH) from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHEMOTAXIS PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Sharma, R, Dhindwal, S, Batra, M, Aggarwal, M, Kumar, P, Tomar, S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pentapeptide-Independent Chemotaxis Receptor Methyltransferase (Cher) Reveals Idiosyncratic Structural Determinants for Receptor Recognition.
J.Struct.Biol., 196, 2016
|
|
8CK1
 
 | | Carin 1 bacteriophage tail, connector and tail fibers assembly | | Descriptor: | Connector Protein, Tail Nozzle, Tail fibers Dpo36 | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
7LSA
 
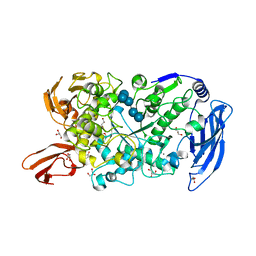 | | Ruminococcus bromii Amy12 with maltoheptaose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
4NA7
 
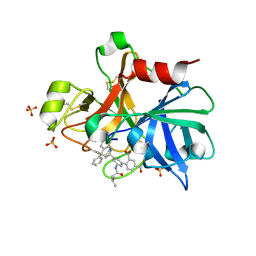 | | Factor XIA in complex with the inhibitor 3'-[(2S,4R)-6-carbamimidoyl-4-methyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]-4-carbamoyl-5'-[(3-methylbutanoyl)amino]biphenyl-2-carboxylic acid | | Descriptor: | 3'-[(2S,4R)-6-carbamimidoyl-4-methyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]-4-carbamoyl-5'-[(3-methylbutanoyl)amino]biphenyl-2-carboxylic acid, Coagulation factor XI, SULFATE ION | | Authors: | Wei, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrahydroquinoline Derivatives as Potent and Selective Factor XIa Inhibitors.
J.Med.Chem., 57, 2014
|
|
7LSR
 
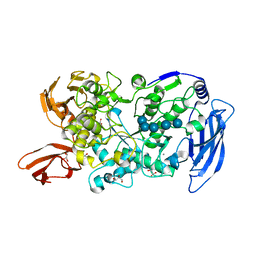 | | Ruminococcus bromii Amy12-D392A with maltoheptaose | | Descriptor: | CALCIUM ION, GLYCEROL, Pullulanase, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
5MC7
 
 | |
4N2R
 
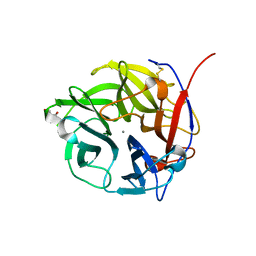 | | Crystal Structure of the alpha-L-arabinofuranosidase UmAbf62A from Ustilago maydis in complex with L-arabinofuranose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, alpha-L-arabinofuranose, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
1KHG
 
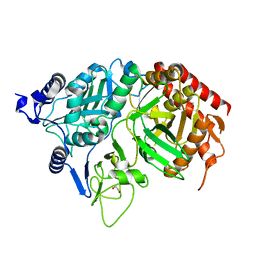 | | PEPCK | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
7ADS
 
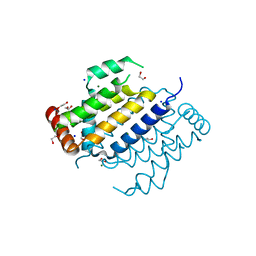 | | Orf virus Apoptosis inhibitor ORFV125 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis inhibitor, CHLORIDE ION, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22032046 Å) | | Cite: | Crystal structures of ORFV125 provide insight into orf virus-mediated inhibition of apoptosis.
Biochem.J., 477, 2020
|
|
8COT
 
 | |
5FNR
 
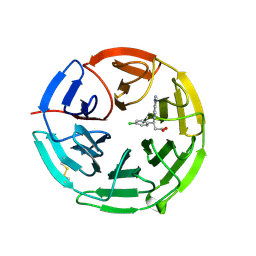 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
7ABJ
 
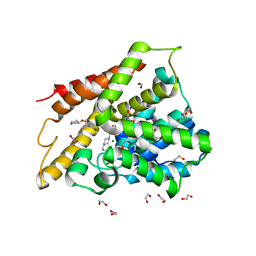 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1361 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(3-(cyclopentyloxy)-4-methoxyphenyl)-2-isopropyl-4,4-dimethyl-2,4-dihydro-3H-pyrazol-3-one, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-1361
To be published
|
|
5FCZ
 
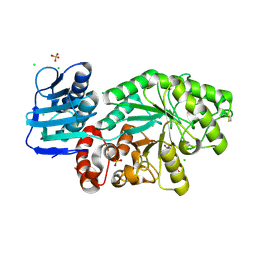 | |
5FE8
 
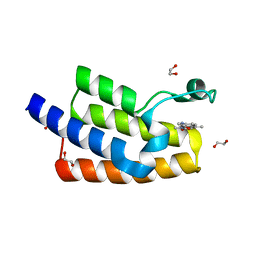 | | Crystal structure of human PCAF bromodomain in complex with compound SL1126 (compound 12) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE4
 
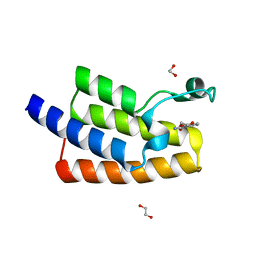 | | Crystal structure of human PCAF bromodomain in complex with fragment MB364 (fragment 5) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1,4-benzodioxine-5-carboxamide, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
1KHF
 
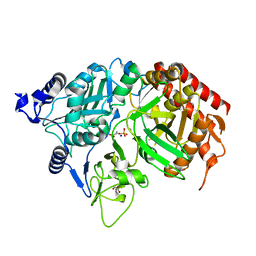 | | PEPCK complex with PEP | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
5FKT
 
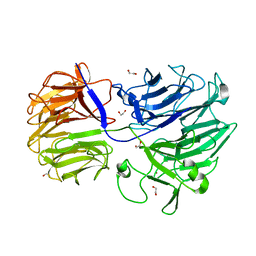 | | Unraveling the first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, ENDO-1,4-BETA-GLUCANASE/XYLOGLUCANASE, ... | | Authors: | Attia, M, Stepper, J, Davies, G.J, Brumer, H. | | Deposit date: | 2015-10-19 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Functional and Structural Characterization of a Potent Gh74 Endo-Xyloglucanase from the Soil Saprophyte Cellvibrio Japonicus Unravels the First Step of Xyloglucan Degradation.
FEBS J., 283, 2016
|
|
7A6S
 
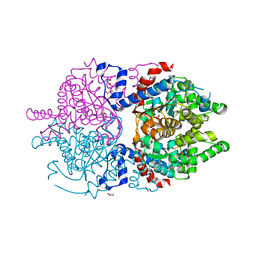 | | Crystal Structure of Asn173Ser variant of Human Deoxyhypusine Synthase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Deoxyhypusine synthase, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-08-26 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
7A6T
 
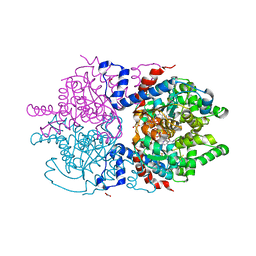 | | Crystal Structure of Asn173Ser variant of Human Deoxyhypusine Synthase in complex with NAD and spermidine | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Deoxyhypusine synthase, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-08-26 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
