4ZN9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Oxabicyclic Heptene Sulfonate (OBHS) | | Descriptor: | Estrogen receptor, Nuclear receptor-interacting peptide, cyclohexa-2,5-dien-1-yl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands.
Chemmedchem, 7, 2012
|
|
1E41
 
 | |
1DFB
 
 | | STRUCTURE OF A HUMAN MONOCLONAL ANTIBODY FAB FRAGMENT AGAINST GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE I | | Descriptor: | IGG1-KAPPA 3D6 FAB (HEAVY CHAIN), IGG1-KAPPA 3D6 FAB (LIGHT CHAIN) | | Authors: | He, X.M, Rueker, F, Casale, E, Carter, D.C. | | Deposit date: | 1992-03-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human monoclonal antibody Fab fragment against gp41 of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1PM6
 
 | | Solution Structure of Full-Length Excisionase (Xis) from Bacteriophage HK022 | | Descriptor: | Excisionase | | Authors: | Rogov, V.V, Luecke, C, Muresanu, L, Wienk, H, Kleinhaus, I, Werner, K, Loehr, F, Pristovsek, P, Rueterjans, H. | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the full-length excisionase from bacteriophage HK022.
Eur.J.Biochem., 270, 2003
|
|
6IDM
 
 | | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Bairagya, H.R, Shokeen, A, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2018-09-10 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution
To Be Published
|
|
6IK9
 
 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
7XY9
 
 | | Cryo-EM structure of secondary alcohol dehydrogenases TbSADH after carrier-free immobilization based on weak intermolecular interactions | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Chen, Q, Li, X, Yang, F, Qu, G, Sun, Z.T, Wang, Y.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Active and stable alcohol dehydrogenase-assembled hydrogels via synergistic bridging of triazoles and metal ions.
Nat Commun, 14, 2023
|
|
1PVG
 
 | |
2CB5
 
 | | HUMAN BLEOMYCIN HYDROLASE, C73S/DELE455 MUTANT | | Descriptor: | PROTEIN (BLEOMYCIN HYDROLASE) | | Authors: | O'Farrell, P.A, Gonzalez, F, Zheng, W, Johnston, S.A, Joshua-Tor, L. | | Deposit date: | 1999-03-02 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human bleomycin hydrolase, a self-compartmentalizing cysteine protease.
Structure Fold.Des., 7, 1999
|
|
5TY3
 
 | | Crystal structure of K72A variant of Human Cytochrome c | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Mou, T.C, Nold, S.M, Lei, H, Sprang, S.R, Bowler, B.E. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effect of a K72A Mutation on the Structure, Stability, Dynamics, and Peroxidase Activity of Human Cytochrome c.
Biochemistry, 56, 2017
|
|
6SWM
 
 | | selenol bound carbonic anhydrase I | | Descriptor: | (2~{R})-1-prop-2-enoxy-3-selanyl-propan-2-ol, Carbonic anhydrase 1, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Selenols: a new class of carbonic anhydrase inhibitors.
Chem.Commun.(Camb.), 55, 2019
|
|
6I9H
 
 | |
6SZH
 
 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GW197 | | Descriptor: | 3,5-dimethyl-1~{H}-pyrrole-2-carbonitrile, CALCIUM ION, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5U5P
 
 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus Musculus Complexed with a MLH1 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
6GHM
 
 | | Structure of PP1 alpha phosphatase bound to ASPP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Apoptosis-stimulating of p53 protein 2, ... | | Authors: | Mouilleron, S, Bertran, T.M, Tapon, N, Zhou, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ASPP proteins discriminate between PP1 catalytic subunits through their SH3 domain and the PP1 C-tail.
Nat Commun, 10, 2019
|
|
5LFN
 
 | | Crystal structure of human chondroadherin | | Descriptor: | CHLORIDE ION, Chondroadherin | | Authors: | Ramisch, S, Pramhed, A, Logan, D.T. | | Deposit date: | 2016-07-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human chondroadherin: solving a difficult molecular-replacement problem using de novo models.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3BOM
 
 | | Crystal structure of trout hemoglobin at 1.35 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Hemoglobin subunit alpha-4, Hemoglobin subunit beta-4, ... | | Authors: | Aranda IV, R, Bingman, C.A, Bitto, E, Wesenberg, G.E, Richards, M, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Trout Hemoglobin Crystal Structure at 1.35 Angstroms Resolution.
To be Published
|
|
2CRT
 
 | |
4O1K
 
 | | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora. | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Lehneck, R, Neumann, P, Vullo, D, Elleuche, S, Supuran, C.T, Ficner, R, Poggeler, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora.
Febs J., 281, 2014
|
|
1UOR
 
 | |
1O3Z
 
 | | HIV-1 DIS(MAL) DUPLEX RU HEXAMINE-SOAKED | | Descriptor: | HIV-1 DIS(MAL) GENOMIC RNA, MAGNESIUM ION, RUTHENIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2003-05-16 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes.
Nucleic Acids Res., 31, 2003
|
|
1MPP
 
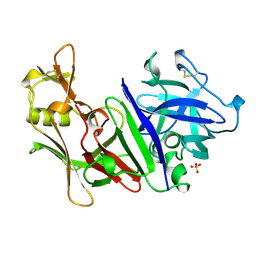 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES. V. STRUCTURE AND REFINEMENT AT 2.0 ANGSTROMS RESOLUTION OF THE ASPARTIC PROTEINASE FROM MUCOR PUSILLUS | | Descriptor: | PEPSIN, SULFATE ION | | Authors: | Newman, M, Watson, F, Roychowdhury, P, Jones, H, Badasso, M, Cleasby, A, Wood, S.P, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of aspartic proteinases. V. Structure and refinement at 2.0 A resolution of the aspartic proteinase from Mucor pusillus.
J.Mol.Biol., 230, 1993
|
|
5T8Q
 
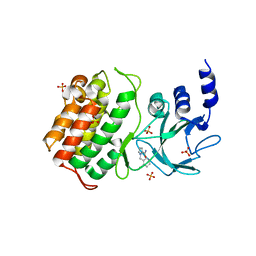 | |
6GUA
 
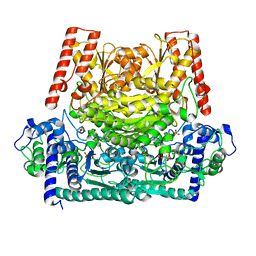 | | Xylulose 5-phosphate phosphoketolase from Lactococcus lactis | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Probable phosphoketolase, ... | | Authors: | Scheidig, A.J, Szedlacsek, S.E. | | Deposit date: | 2018-06-19 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | SOLUTION SCATTERING (1.95 Å), X-RAY DIFFRACTION | | Cite: | Crystal structure of a xylulose 5-phosphate phosphoketolase. Insights into the substrate specificity for xylulose 5-phosphate.
J.Struct.Biol., 207, 2019
|
|
6GH7
 
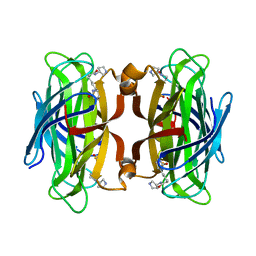 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[(3~{R})-pyrrolidin-3-yl]pentanamide, Streptavidin | | Authors: | Nodling, A.R, Tsai, Y.H, Luk, L.Y.P, Rizkallah, P, Jin, Y. | | Deposit date: | 2018-05-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Reactivity and Selectivity of Iminium Organocatalysis Improved by a Protein Host.
Angew.Chem.Int.Ed.Engl., 57, 2018
|
|
