3RQ3
 
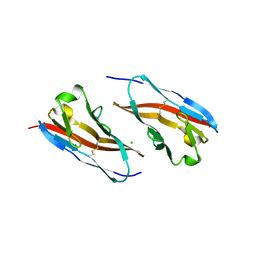 | | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form | | Descriptor: | CHLORIDE ION, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Ramagopal, U.A, Rubinstein, R, Guo, H, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-04-27 | | Release date: | 2011-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form
To be published
|
|
3RRQ
 
 | |
3Q0H
 
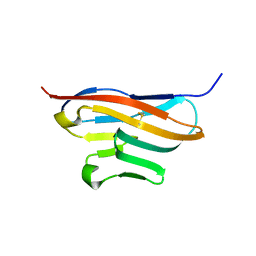 | | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) | | Descriptor: | T cell immunoreceptor with Ig and ITIM domains | | Authors: | Ramagopal, U.A, Guo, H, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT)
To be published
|
|
6IGO
 
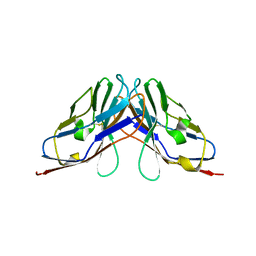 | | Crystal structure of myelin protein zero-like protein 1 (MPZL1) | | Descriptor: | Myelin protein zero-like protein 1 | | Authors: | Yu, T. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6ISA
 
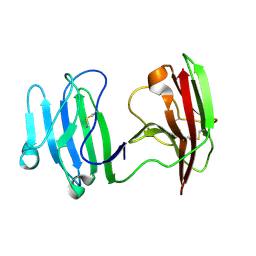 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ISB
 
 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6J14
 
 | | Complex structure of GY-14 and PD-1 | | Descriptor: | GY-14 heavy chain V fragment, GY-14 light chain V fragment, Programmed cell death protein 1 | | Authors: | Chen, D, Tan, S, Whang, H, Zhang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2018-12-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6HIG
 
 | | hPD-1/NBO1a Fab complex | | Descriptor: | Heavy Chain, Light Chain, Programmed cell death protein 1 | | Authors: | Loredo-Varela, J.L, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2018-08-29 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tumor suppression of novel anti-PD-1 antibodies mediated through CD28 costimulatory pathway.
J.Exp.Med., 216, 2019
|
|
6JJP
 
 | | Crystal structure of Fab of a PD-1 monoclonal antibody MW11-h317 in complex with PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of MW11-h317, Programmed cell death protein 1, ... | | Authors: | Wang, M, Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M. | | Deposit date: | 2019-02-26 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation.
Commun Biol, 2, 2019
|
|
6K0Y
 
 | | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Liu, J.X, Wang, G.Q. | | Deposit date: | 2019-05-08 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor.
Sci Rep, 9, 2019
|
|
8GY5
 
 | |
8GAB
 
 | | Crystal structure of CTLA-4 in complex with a high affinity CTLA-4 binder | | Descriptor: | CTLA-4 binder, Cytotoxic T-lymphocyte protein 4, POTASSIUM ION | | Authors: | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
6ISC
 
 | | complex structure of mCD226-ecto and hCD155-D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, Poliovirus receptor | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6IGT
 
 | | MPZL1 mutant - V145G, Q146K, P147T and G148S | | Descriptor: | Myelin protein zero-like protein 1 | | Authors: | Yu, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IGW
 
 | | MPZL1 mutant - S86G, V145G, Q146K,P147T,G148S | | Descriptor: | Myelin protein zero-like protein 1 | | Authors: | Yu, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
8HIT
 
 | | Crystal structure of anti-CTLA-4 humanized IgG1 MAb--JS007 in complex with human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, JS007-VH, JS007-VL | | Authors: | Tan, S, Shi, Y, Wang, Q, Gao, G.F, Guan, J, Chai, Y, Qi, J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of the high-affinity anti-CTLA-4 monoclonal antibody JS007 for immune checkpoint therapy of cancer.
Mabs, 15, 2023
|
|
8HXS
 
 | | Small_spotted catshark CD8alpha | | Descriptor: | T-cell surface glycoprotein CD8 alpha chain | | Authors: | Wang, J, Zou, J. | | Deposit date: | 2023-01-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The first crystal structure of CD8 alpha alpha from a cartilaginous fish.
Front Immunol, 14, 2023
|
|
6J15
 
 | | Complex structure of GY-5 Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GY-5 heavy chain Fab, ... | | Authors: | Chen, D, Tan, S, Zhang, H, Wang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2018-12-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
6JBT
 
 | | Complex structure of toripalimab-Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Guo, L, Tan, S, Chai, Y, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy.
Mabs, 11, 2019
|
|
3DUS
 
 | | Crystal structure of SAG506-01, orthorhombic, twinned, crystal 1 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3DV4
 
 | | Crystal structure of SAG506-01, tetragonal, crystal 1 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-18 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3DUR
 
 | | Crystal structure of SAG173-04 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3DUU
 
 | | Crystal structure of SAG506-01, orthorhombic, twinned, crystal 2 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3DV6
 
 | | Crystal structure of SAG506-01, tetragonal, crystal 2 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-18 | | Release date: | 2008-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
