2FVR
 
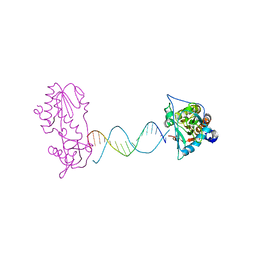 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*TP*CP*TP*TP*TP*CP*AP*TP*AP*TP*GP*AP*AP*AP*GP*A)-3', reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
1N1Q
 
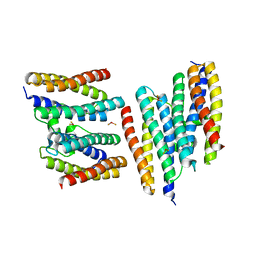 | | Crystal structure of a Dps protein from Bacillus brevis | | Descriptor: | DPS Protein, MU-OXO-DIIRON | | Authors: | Ren, B, Tibbelin, G, Kajino, T, Asami, O, Ladenstein, R. | | Deposit date: | 2002-10-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Multi-layered Structure of Dps with a Novel Di-nuclear Ferroxidase Center
J.Mol.Biol., 329, 2003
|
|
2FVQ
 
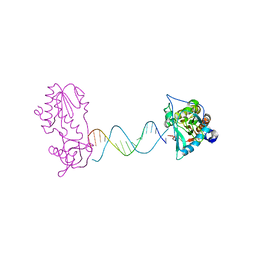 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*CP*TP*TP*TP*CP*AP*TP*TP*AP*AP*TP*GP*AP*AP*AP*G)-3', reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
2FVP
 
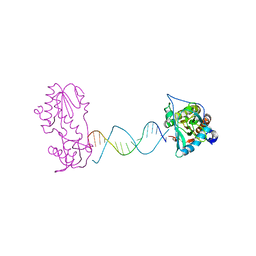 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*TP*GP*CP*AP*AP*TP*GP*AP*AP*A)-3', Reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
1BBO
 
 | |
2K29
 
 | |
4E89
 
 | | Crystal Structure of RnaseH from gammaretrovirus | | Descriptor: | CADMIUM ION, MAGNESIUM ION, RNase H | | Authors: | Kim, J.H, Kim, S.J. | | Deposit date: | 2012-03-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of xenotropic murine leukaemia virus-related virus (XMRV) ribonuclease H
Biosci.Rep., 32, 2012
|
|
2FVS
 
 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*CP*AP*CP*AP*AP*TP*GP*AP*TP*CP*AP*TP*TP*GP*TP*G)-3', reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
4CIJ
 
 | |
1GHC
 
 | | HOMO-AND HETERONUCLEAR TWO-DIMENSIONAL NMR STUDIES OF THE GLOBULAR DOMAIN OF HISTONE H1: FULL ASSIGNMENT, TERTIARY STRUCTURE, AND COMPARISON WITH THE GLOBULAR DOMAIN OF HISTONE H5 | | Descriptor: | GH1 | | Authors: | Cerf, C, Lippens, G, Ramakrishnan, V, Muyldermans, S, Segers, A, Wyns, L, Wodak, S.J, Hallenga, K. | | Deposit date: | 1994-05-16 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure, and comparison with the globular domain of histone H5.
Biochemistry, 33, 1994
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1HHX
 
 | | Solution structure of LNA3:RNA hybrid | | Descriptor: | 5- D(*CP*+TP*GP*AP*+TP*AP*+TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
4CWE
 
 | |
1LW0
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
2G1Z
 
 | |
2GDO
 
 | | 4-(Aminoalkylamino)-3-Benzimidazole-Quinolinones As Potent CHK1 Inhibitors | | Descriptor: | 4-[(3S)-1-AZABICYCLO[2.2.2]OCT-3-YLAMINO]-3-(1H-BENZIMIDAZOL-2-YL)-6-CHLOROQUINOLIN-2(1H)-ONE, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Le, V, Dove, J, Fang, E, Bussiere, D.E. | | Deposit date: | 2006-03-16 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4-(Aminoalkylamino)-3-benzimidazole-quinolinones as potent CHK-1 inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2LUA
 
 | | Solution structure of CXC domain of MSL2 | | Descriptor: | Protein male-specific lethal-2, ZINC ION | | Authors: | Feng, Y, Ye, K, Zheng, S, Wang, J. | | Deposit date: | 2012-06-09 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of MSL2 CXC Domain Reveals an Unusual Zn(3)Cys(9) Cluster and Similarity to Pre-SET Domains of Histone Lysine Methyltransferases.
Plos One, 7, 2012
|
|
3QP4
 
 | | Crystal structure of CviR ligand-binding domain bound to C10-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QPH
 
 | | The three-dimensional structure of TrmB, a global transcriptional regulator of the hyperthermophilic archaeon Pyrococcus furiosus in complex with sucrose | | Descriptor: | ACETATE ION, GLYCEROL, TrmB, ... | | Authors: | Krug, M, Lee, S.-J, Boos, W, Welte, W, Diederichs, K. | | Deposit date: | 2011-02-13 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | The three-dimensional structure of TrmB, a transcriptional regulator of dual function in the hyperthermophilic archaeon Pyrococcus furiosus in complex with sucrose.
Protein Sci., 22, 2013
|
|
6Z8W
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3G), which contains 1-beta-D-glucopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
3QP2
 
 | | Crystal structure of CviR ligand-binding domain bound to C8-HSL | | Descriptor: | CviR transcriptional regulator, N-(2-OXOTETRAHYDROFURAN-3-YL)OCTANAMIDE | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP8
 
 | | Crystal structure of CviR (Chromobacterium violaceum 12472) ligand-binding domain bound to C10-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
6Z8X
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3Leu), which contains leucyl amide in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
4ZNJ
 
 | | Thermus Phage P74-26 Large Terminase ATPase domain mutant R139A (I 2 3 space group) | | Descriptor: | Phage terminase large subunit, SULFATE ION | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6Z8V
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3L), which contains 1-beta-D-lactopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
