8OFE
 
 | | E.coli Peptide Deformylase with bound inhibitor | | Descriptor: | (2R)-2-[[methanoyl(oxidanyl)amino]methyl]-N-[(2S)-3-methyl-1-oxidanylidene-1-[2-(sulfanylmethyl)pyrrolidin-1-yl]butan-2-yl]heptanamide, Peptide deformylase, ZINC ION | | Authors: | Kirschner, H, Stoll, R, Hofmann, E. | | Deposit date: | 2023-03-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights into Antibacterial Payload Release from Gold Nanoparticles Bound to E. coli Peptide Deformylase.
Chemmedchem, 19, 2024
|
|
7ALJ
 
 | | Structure of Drosophila Notch EGF domains 11-13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurogenic locus Notch protein, ... | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-06 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
7ALK
 
 | | Structure of Drosophila C2-DSL-EGF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neurogenic locus protein delta, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-06 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
7ALT
 
 | | Structure of Drosophila Serrate C2-DSL-EGF1-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
6V3P
 
 | |
4WDY
 
 | | JC Polyomavirus VP1 five-fold pore mutant N221Q | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1 | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a Pore in the Capsid of JC Polyomavirus Reduces Infectivity and Prevents Exposure of the Minor Capsid Proteins.
J.Virol., 89, 2015
|
|
5LSG
 
 | | PPARgamma complex with the betulinic acid | | Descriptor: | Betulinic Acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Calleri, E, Paiardini, A. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Betulinic acid is a PPAR gamma antagonist that improves glucose uptake, promotes osteogenesis and inhibits adipogenesis.
Sci Rep, 7, 2017
|
|
1K6L
 
 | | Photosynethetic Reaction Center from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Pokkuluri, P.R, Laible, P.D, Deng, Y.-L, Wong, T.N, Hanson, D.K, Schiffer, M. | | Deposit date: | 2001-10-16 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of a mutant photosynthetic reaction center shows unexpected changes in main chain orientations and quinone position.
Biochemistry, 41, 2002
|
|
1K6N
 
 | | E(L212)A,D(L213)A Double Mutant Structure of Photosynthetic Reaction Center from Rhodobacter Sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Pokkuluri, P.R, Laible, P.D, Deng, Y.-L, Wong, T.N, Hanson, D.K, Schiffer, M. | | Deposit date: | 2001-10-16 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of a mutant photosynthetic reaction center shows unexpected changes in main chain orientations and quinone position.
Biochemistry, 41, 2002
|
|
1YAE
 
 | | Structure of the Kainate Receptor Subunit GluR6 Agonist Binding Domain Complexed with Domoic Acid | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor, ... | | Authors: | Nanao, M.H, Green, T, Stern-Bach, Y, Heinemann, S.F, Choe, S. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure of the kainate receptor subunit GluR6 agonist-binding domain complexed with domoic acid.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7STA
 
 | |
8BXB
 
 | |
1KE0
 
 | |
5ZVC
 
 | | P domain of GII.13 norovirus capsid complexed with Lewis A trisaccharide | | Descriptor: | GLYCEROL, Major capsid protein VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
3GXU
 
 | | Crystal structure of Eph receptor and ephrin complex | | Descriptor: | Ephrin type-A receptor 4, Ephrin-B2 | | Authors: | Qin, H.N, Song, J.X. | | Deposit date: | 2009-04-03 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of the EphA4-ephrin-B2 complex reveals new features enabling Eph-ephrin binding promiscuity
J.Biol.Chem., 2009
|
|
5ZV9
 
 | | P domain of GII.13 norovirus capsid | | Descriptor: | GLYCEROL, Major capsid protein VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
1KE3
 
 | |
5W87
 
 | |
1Y26
 
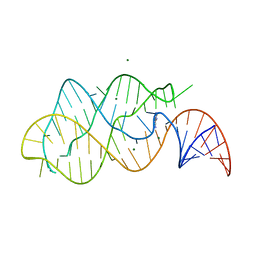 | | A-riboswitch-adenine complex | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus A-riboswitch | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
1KDW
 
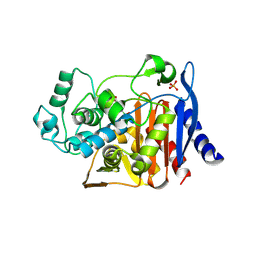 | |
1KDS
 
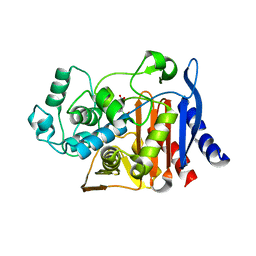 | |
8QMO
 
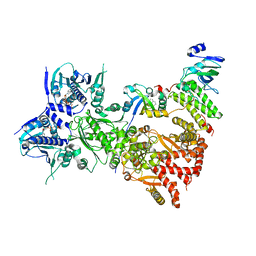 | | Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Kwong, H.S, Grandvuillemin, L, Sirounian, S, Ancelin, A, Lai-Kee-Him, J, Carivenc, C, Lancey, C, Ragan, T.J, Hesketh, E.L, Bourguet, W, Gruszczyk, J. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Insights into the Activation of Human Aryl Hydrocarbon Receptor by the Environmental Contaminant Benzo[a]pyrene and Structurally Related Compounds.
J.Mol.Biol., 436, 2024
|
|
3HY4
 
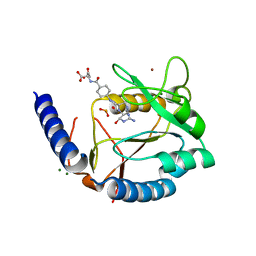 | | Structure of human MTHFS with N5-iminium phosphate | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, N-({trans-4-[({(2R,4R,4aS,6S,8aS)-2-amino-4-hydroxy-5-[(phosphonooxy)methyl]decahydropteridin-6-yl}methyl)amino]cyclohexyl}carbonyl)-L-glutamic acid, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
1AXY
 
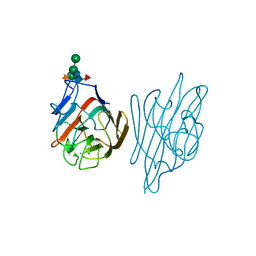 | | ERYTHRINA CORALLODENDRON LECTIN | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Shaanan, B, Elgavish, S. | | Deposit date: | 1997-10-24 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Erythrina corallodendron lectin and of its complexes with mono- and disaccharides.
J.Mol.Biol., 277, 1998
|
|
1AX2
 
 | |
