7UF7
 
 | |
2HFH
 
 | |
7UF6
 
 | |
2GLT
 
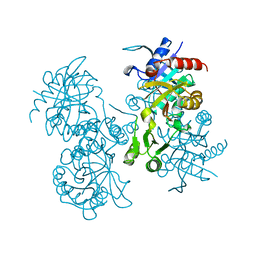 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
2HCO
 
 | |
2GTU
 
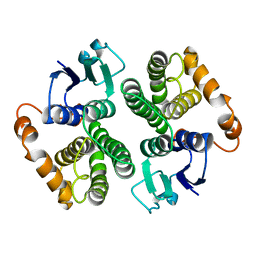 | | LIGAND-FREE HUMAN GLUTATHIONE S-TRANSFERASE M2-2 (E.C.2.5.1.18), MONOCLINIC CRYSTAL FORM | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Patskovska, L.N, Fedorov, A.A, Patskovsky, Y.V, Almo, S.C, Listowsky, I. | | Deposit date: | 1998-05-26 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The enhanced affinity for thiolate anion and activation of enzyme-bound glutathione is governed by an arginine residue of human Mu class glutathione S-transferases.
J.Biol.Chem., 275, 2000
|
|
2GSQ
 
 | |
2HAD
 
 | |
2HBS
 
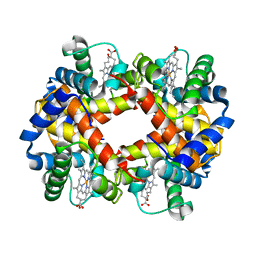 | | THE HIGH RESOLUTION CRYSTAL STRUCTURE OF DEOXYHEMOGLOBIN S | | Descriptor: | HEMOGLOBIN S (DEOXY), ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Harrington, D.J, Adachi, K, Royer Junior, W.E. | | Deposit date: | 1997-05-06 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The high resolution crystal structure of deoxyhemoglobin S.
J.Mol.Biol., 272, 1997
|
|
2DMR
 
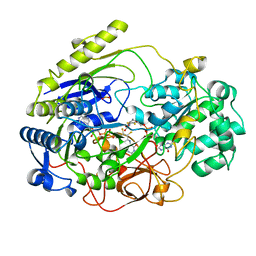 | | DITHIONITE REDUCED DMSO REDUCTASE FROM RHODOBACTER CAPSULATUS | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM(IV) ION, ... | | Authors: | Mcalpine, A.S, Bailey, S. | | Deposit date: | 1997-04-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molybdenum Active Centre of Dmso Reductase from Rhodobacter Capsulatus: Crystal Structure of the Oxidised Enzyme at 1.82-A Resolution and the Dithionite-Reduced Enzyme at 2.8-A Resolution
J.Biol.Inorg.Chem., 2, 1997
|
|
2DAP
 
 | | C. GLUTAMICUM DAP DEHYDROGENASE IN COMPLEX WITH DAP | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, DIAMINOPIMELIC ACID DEHYDROGENASE | | Authors: | Scapin, G, Cirilli, M, Reddy, S.G, Gao, Y, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 1997-12-23 | | Release date: | 1998-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate and inhibitor binding sites in Corynebacterium glutamicum diaminopimelate dehydrogenase.
Biochemistry, 37, 1998
|
|
7U6Z
 
 | | Pertussis toxin E129D NAD | | Descriptor: | IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2022-03-06 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.30002 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7UB0
 
 | |
2HKK
 
 | | Carbonic anhydrase activators: Solution and X-ray crystallography for the interaction of andrenaline with various carbonic anhydrase isoforms | | Descriptor: | Carbonic anhydrase 2, L-EPINEPHRINE, MERCURY (II) ION, ... | | Authors: | Temperini, C, Innocenti, A, Vullo, D, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2006-07-05 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase activators: L-Adrenaline plugs the active site entrance of isozyme II, activating better isoforms I, IV, VA, VII, and XIV.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7UB6
 
 | |
7UB5
 
 | |
2E3Z
 
 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in substrate-free form | | Descriptor: | Beta-glucosidase | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
2KBC
 
 | |
2KCF
 
 | |
2KFY
 
 | |
7UTI
 
 | | ALTERNATIVE MODELING OF TROPOMYOSIN IN HUMAN CARDIAC THIN FILAMENT IN THE CALCIUM BOUND STATE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rynkiewicz, M.J, Pavadai, E, Lehman, W. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Protein-Protein Docking Reveals Dynamic Interactions of Tropomyosin on Actin Filaments.
Biophys J, 119, 2020
|
|
2KIU
 
 | |
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
7UTL
 
 | | ALTERNATIVE MODELING OF TROPOMYOSIN IN HUMAN CARDIAC THIN FILAMENT IN THE CALCIUM FREE STATE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rynkiewicz, M.J, Pavadai, E, Lehman, W. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Protein-Protein Docking Reveals Dynamic Interactions of Tropomyosin on Actin Filaments.
Biophys J, 119, 2020
|
|
2E99
 
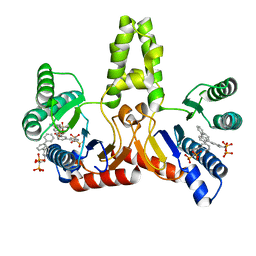 | | E. coli undecaprenyl pyrophosphate synthase in complex with BPH-608 | | Descriptor: | (1-HYDROXY-1-PHOSPHONO-2-[1,1';3',1'']TERPHENYL-3-YL-ETHYL)-PHOSPHONIC ACID, Undecaprenyl pyrophosphate synthetase | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Liang, P.H, Oldfield, E, Wang, A.H.J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
