8QW2
 
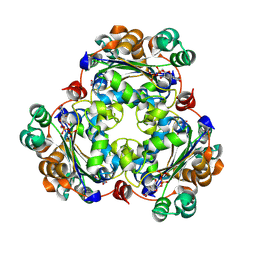 | | Human NDPK-C in complex with UDP and Mg2+ | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase 3, URIDINE-5'-DIPHOSPHATE | | Authors: | Werten, S, Amjadi, R, Scheffzek, K. | | Deposit date: | 2023-10-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanistic Insights into Substrate Recognition of Human Nucleoside Diphosphate Kinase C Based on Nucleotide-Induced Structural Changes.
Int J Mol Sci, 25, 2024
|
|
8QW0
 
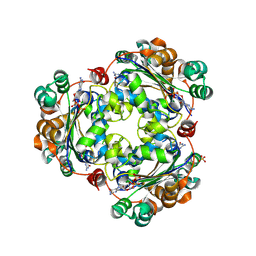 | | Human NDPK-C in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase 3, SULFATE ION | | Authors: | Werten, S, Amjadi, R, Scheffzek, K. | | Deposit date: | 2023-10-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Mechanistic Insights into Substrate Recognition of Human Nucleoside Diphosphate Kinase C Based on Nucleotide-Induced Structural Changes.
Int J Mol Sci, 25, 2024
|
|
8QP6
 
 | |
8R0C
 
 | |
8QPQ
 
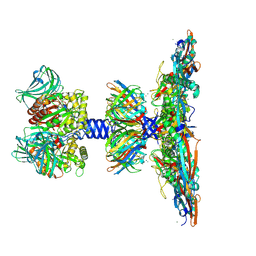 | | C1 turret to capsid interface of full Haloferax tailed virus 1 adjacent to the portal-capsid interface. | | Descriptor: | HK97 gp5-like major capsid protein, MAGNESIUM ION, Prokaryotic polysaccharide deacetylase, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
6SKO
 
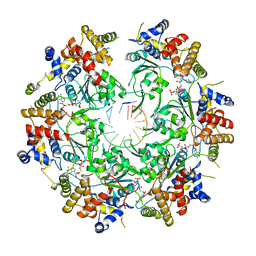 | | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNA | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
8QWE
 
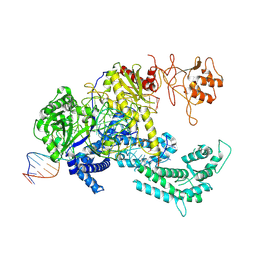 | |
6SKN
 
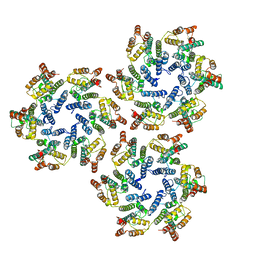 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-26 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8OHZ
 
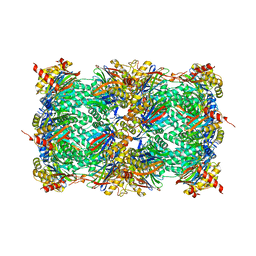 | | Yeast 20S proteasome in complex with a photoswitchable cepafungin derivative (transCep1) | | Descriptor: | (2~{S},3~{R})-2-[2-[4-[2-(4-ethylphenyl)hydrazinyl]phenyl]ethanoylamino]-~{N}-[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]-3-oxidanyl-butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Morstein, J, Amatuni, A, Schuster, A, Kuttenlochner, W, Ko, T, Groll, M, Adibekian, A, Renata, H, Trauner, D.H. | | Deposit date: | 2023-03-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optical Control of Proteasomal Protein Degradation with a Photoswitchable Lipopeptide.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QBX
 
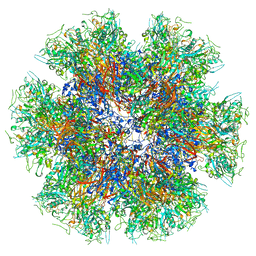 | | Chimeric Adenovirus-derived dodecamer | | Descriptor: | Penton protein | | Authors: | Buzas, D, Borucu, U, Bufton, J, Kapadalakere, S.Y, Toelzer, C. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8Q7Y
 
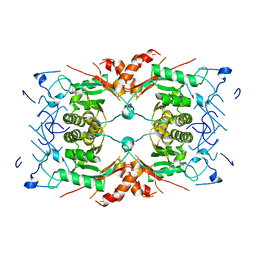 | |
8QCF
 
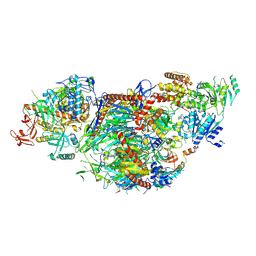 | | yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Antiviral helicase SKI2, Exosome complex component CSL4, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
6SJJ
 
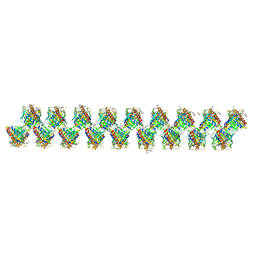 | | A new modulated crystal structure of ANS complex of St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, CITRATE ANION, ... | | Authors: | Smietanska, J, Sliwiak, J, Gilski, M, Dauter, Z, Strzalka, R, Wolny, J, Jaskolski, M. | | Deposit date: | 2019-08-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new modulated crystal structure of the ANS complex of the St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8QY9
 
 | |
6SKP
 
 | |
8QYB
 
 | |
6SZI
 
 | | ASR Alternansucrase in complex with isomaltose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
6S8O
 
 | | Human Brr2 Helicase Region M641C/A1582C | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Vester, K, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-07-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.172 Å) | | Cite: | The inactive C-terminal cassette of the dual-cassette RNA helicase BRR2 both stimulates and inhibits the activity of the N-terminal helicase unit.
J.Biol.Chem., 295, 2020
|
|
6T16
 
 | | ASR Alternansucrase in complex with panose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
8QJS
 
 | | VHL/Elongin B/Elongin C complex with compound 155 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-(2-methoxyethoxy)ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
6SKR
 
 | | OXA-10_ETP. Structural insight to the enhanced carbapenem efficiency of OXA-655 compared to OXA-10. | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, FORMIC ACID, ... | | Authors: | Leiros, H.-K.S. | | Deposit date: | 2019-08-16 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the enhanced carbapenemase efficiency of OXA-655 compared to OXA-10.
Febs Open Bio, 10, 2020
|
|
3QP3
 
 | |
6QXP
 
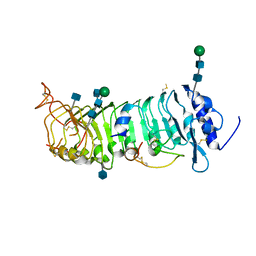 | | Protein peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat extensin-like protein 2, ... | | Authors: | Moussu, S, Caroline, C, Santos-Fernandez, G, Wehrle, S, Grossniklaus, U, Santiago, J. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QWS
 
 | |
6QQ7
 
 | |
