1E5N
 
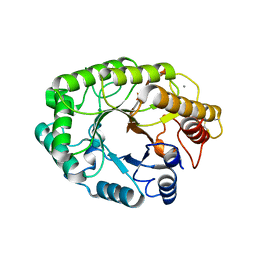 | | E246C mutant of P fluorescens subsp. cellulosa xylanase A in complex with xylopentaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Lo Leggio, L, Jenkins, J.A, Harris, G.W, Pickersgill, R.W. | | Deposit date: | 2000-07-27 | | Release date: | 2000-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic study of xylopentaose binding to Pseudomonas fluorescens xylanase A.
Proteins, 41, 2000
|
|
1HY2
 
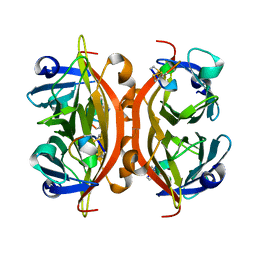 | | MINIPROTEIN MP-1 COMPLEX WITH STREPTAVIDIN | | Descriptor: | MP-1, STREPTAVIDIN | | Authors: | Yang, H.W, Liu, D.Q, Fan, X, White, M.A, Fox, R.O. | | Deposit date: | 2001-01-17 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Ensemble Analysis of Ligand Binding in Streptavidin Mini-Protein Complexes
To be Published
|
|
1E6Z
 
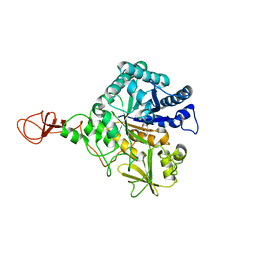 | | CHITINASE B FROM SERRATIA MARCESCENS WILDTYPE IN COMPLEX WITH CATALYTIC INTERMEDIATE | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-23 | | Release date: | 2001-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HXL
 
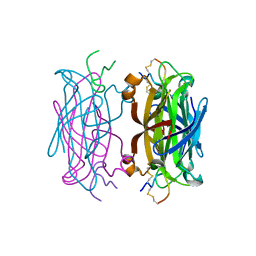 | | MINIPROTEIN MP-2 (V10A) COMPLEX WITH STREPTAVIDIN | | Descriptor: | MP-2, STREPTAVIDIN | | Authors: | Yang, H.W, Liu, D.Q, Fan, X, White, M.A, Fox, R.O. | | Deposit date: | 2001-01-16 | | Release date: | 2003-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Ensemble Analysis of Ligand Binding in Streptavidin Mini-Protein Complexes
To be Published
|
|
1I08
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE H30A MUTANT OF MANGANESE SUPEROXIDE DISMUTASE FROM E. COLI | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2001-01-29 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Removing a hydrogen bond in the dimer interface of Escherichia coli manganese superoxide dismutase alters structure and reactivity.
Biochemistry, 40, 2001
|
|
1HXZ
 
 | | MINIPROTEIN MP-2 COMPLEX WITH STREPTAVIDIN | | Descriptor: | MP-2, STREPTAVIDIN | | Authors: | Yang, H.W, Liu, D.Q, Fan, X, White, M.A, Fox, R.O. | | Deposit date: | 2001-01-17 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Ensemble Analysis of Ligand Binding in Streptavidin Mini-Protein Complexes
To be Published
|
|
1ELY
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102C) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
1EV2
 
 | | CRYSTAL STRUCTURE OF FGF2 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 2 (FGFR2) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 2), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 2), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1EXU
 
 | | CRYSTAL STRUCTURE OF THE HUMAN MHC-RELATED FC RECEPTOR | | Descriptor: | BETA-2-MICROGLOBULIN, BETA-MERCAPTOETHANOL, IGG RECEPTOR FCRN LARGE SUBUNIT P51 | | Authors: | West Jr, A.P, Bjorkman, P.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-08-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and immunoglobulin G binding properties of the human major histocompatibility complex-related Fc receptor(,).
Biochemistry, 39, 2000
|
|
4D41
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-hexyl- 2-(4-nitrophenoxy)phenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5-HEXYL-2-(4-NITROPHENOXY)PHENOL, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
1EV5
 
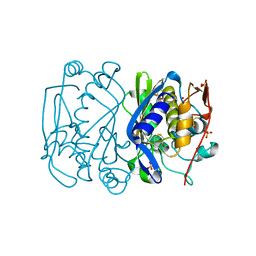 | | CRYSTAL STRUCTURE ANALYSIS OF ALA167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1IA7
 
 | | CRYSTAL STRUCTURE OF THE CELLULASE CEL9M OF C. CELLULOLYTICIUM IN COMPLEX WITH CELLOBIOSE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULASE CEL9M, ... | | Authors: | Parsiegla, G, Belaich, A, Belaich, J.P, Haser, R. | | Deposit date: | 2001-03-22 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the cellulase Cel9M enlightens structure/function relationships of the variable catalytic modules in glycoside hydrolases.
Biochemistry, 41, 2002
|
|
1EVG
 
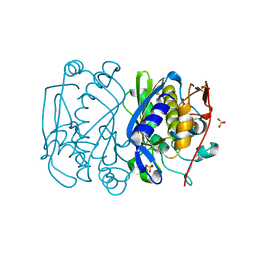 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI WITH UNMODIFIED CATALYTIC CYSTEINE | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1IB6
 
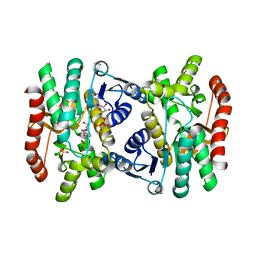 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-03-27 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
1I0S
 
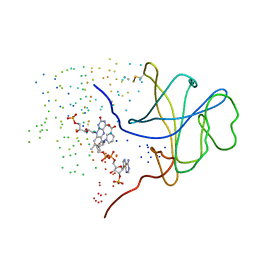 | | ARCHAEOGLOBUS FULGIDUS FERRIC REDUCTASE COMPLEX WITH NADP+ | | Descriptor: | CONSERVED HYPOTHETICAL PROTEIN, FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Chiu, H.-J, Johnson, E, Schroder, I, Rees, D.C. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus and its complex with NADP+.
Structure, 9, 2001
|
|
1I0R
 
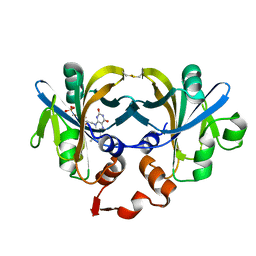 | | CRYSTAL STRUCTURE OF FERRIC REDUCTASE FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | CONSERVED HYPOTHETICAL PROTEIN, FLAVIN MONONUCLEOTIDE | | Authors: | Chiu, H.-J, Johnson, E, Schroder, I, Rees, D.C. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-02 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus and its complex with NADP+.
Structure, 9, 2001
|
|
1EXN
 
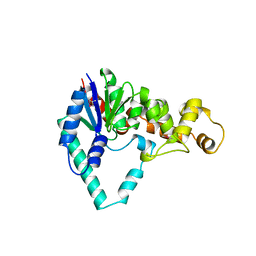 | | T5 5'-EXONUCLEASE | | Descriptor: | 5'-EXONUCLEASE | | Authors: | Ceska, T.A, Sayers, J.R, Stier, G, Suck, D. | | Deposit date: | 1997-01-17 | | Release date: | 1997-07-07 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A helical arch allowing single-stranded DNA to thread through T5 5'-exonuclease.
Nature, 382, 1996
|
|
1EVT
 
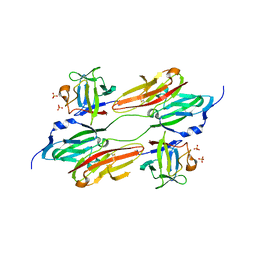 | | CRYSTAL STRUCTURE OF FGF1 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 1 (FGFR1) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 1), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 1), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1I4P
 
 | |
1I1Q
 
 | |
1I4W
 
 | | THE CRYSTAL STRUCTURE OF THE TRANSCRIPTION FACTOR SC-MTTFB OFFERS INTRIGUING INSIGHTS INTO MITOCHONDRIAL TRANSCRIPTION | | Descriptor: | MITOCHONDRIAL REPLICATION PROTEIN MTF1, XENON | | Authors: | Schubot, F.D, Chen, C.-J, Rose, J.P, Dailey, T.A, Dailey, H.A, Wang, B.-C. | | Deposit date: | 2001-02-23 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription.
Protein Sci., 10, 2001
|
|
1EXT
 
 | |
1EVE
 
 | | THREE DIMENSIONAL STRUCTURE OF THE ANTI-ALZHEIMER DRUG, E2020 (ARICEPT), COMPLEXED WITH ITS TARGET ACETYLCHOLINESTERASE | | Descriptor: | 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kryger, G, Silman, I, Sussman, J.L. | | Deposit date: | 1998-03-04 | | Release date: | 1999-01-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of acetylcholinesterase complexed with E2020 (Aricept): implications for the design of new anti-Alzheimer drugs.
Structure Fold.Des., 7, 1999
|
|
1I8L
 
 | | HUMAN B7-1/CTLA-4 CO-STIMULATORY COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOTOXIC T-LYMPHOCYTE PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD80, ... | | Authors: | Stamper, C.C, Somers, W.S, Mosyak, L. | | Deposit date: | 2001-03-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses.
Nature, 410, 2001
|
|
1EMF
 
 | |
