4I5F
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I9N
 
 | | Crystal structure of rabbit LDHA in complex with AP28161 and AP28122 | | Descriptor: | 6-({2-[(5-chloro-4-{[(2S)-2,3-dihydroxypropyl]oxy}-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, 6-[3-(carboxymethoxy)-5-fluorophenyl]pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4HZ8
 
 | | Crystal structure of BglB with natural substrate | | Descriptor: | Beta-glucosidase, beta-D-glucopyranose | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural insights into the substrate recognition properties of beta-glucosidase.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
4HZ7
 
 | | Crystal structure of BglB with glucose | | Descriptor: | beta-D-glucopyranose, beta-glucosidase | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate recognition properties of beta-glucosidase.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
4I3A
 
 | | Structures of PR1 and PR2 intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4I8G
 
 | | Bovine trypsin at 0.8 resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J9P
 
 | | Human DNA polymerase eta-DNA postinsertion binary complex with TA base pair | | Descriptor: | DNA (5'-D(*C*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*TP*A)-3'), DNA polymerase eta, ... | | Authors: | Zhao, Y, Gregory, M, Biertumpfel, C, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2013-02-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of somatic hypermutation at the WA motif by human DNA polymerase eta.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JCI
 
 | | Crystal structure of csal_2705, a putative hydroxyproline epimerase from CHROMOHALOBACTER SALEXIGENS (TARGET EFI-506486), SPACE GROUP P212121, unliganded | | Descriptor: | Proline racemase, SODIUM ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of csal_2705, a putative hydroxyproline epimerase from CHROMOHALOBACTER SALEXIGENS (TARGET EFI-506486), space group P212121, unliganded
To be Published
|
|
4JVV
 
 | | Crystal structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1, covalently bound with diisopropyl fluorophosphate (DFP), Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | evolved variant of the computationally designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4JQJ
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Aminoquinoline | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JGK
 
 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275 | | Descriptor: | evolved variant of a designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275
To be Published
|
|
4JUZ
 
 | | Ternary complex of gamma-OHPDG adduct modified dna (zero primer) with dna polymerase iv and incoming dgtp | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
4JLP
 
 | |
4JM8
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2,6-diaminopyridine | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JMV
 
 | |
4G57
 
 | | Staphylococcal Nuclease double mutant I72L, I92L | | Descriptor: | Thermonuclease | | Authors: | Sakon, J, Stites, W.E, Gill, E, Latimer, E, Rosser, J. | | Deposit date: | 2012-07-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: |
|
|
4FUE
 
 | | Crystal Structure of the Urokinase | | Descriptor: | 6-(1,2,3,4-tetrahydroisoquinolin-6-ylethynyl)naphthalene-2-carboximidamide, ACETATE ION, GLYCEROL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Nienaber, V, Giranda, V. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Urokinase
to be published
|
|
4FTX
 
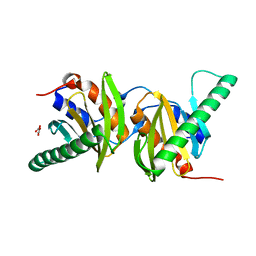 | | Crystal structure of Ego3 homodimer | | Descriptor: | Protein SLM4, SUCCINIC ACID | | Authors: | Zhang, T, Peli-Gulli, M.P, Yang, H, De Virgilio, C, Ding, J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Structure, 20, 2012
|
|
4FUG
 
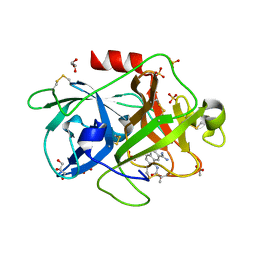 | | Crystal Structure of the Urokinase | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Nienaber, V, Giranda, V. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Urokinase
to be published
|
|
4ILE
 
 | |
4IIQ
 
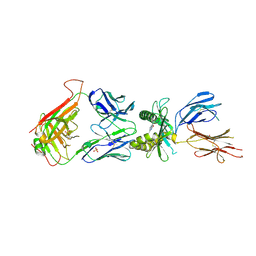 | | Crystal structure of a human MAIT TCR in complex with bovine MR1 | | Descriptor: | Beta-2-microglobulin, MHC class I-related protein, Human Mucosal Associated Invariant T cell receptor alpha chain, ... | | Authors: | Lopez-Sagaseta, J, Adams, E.J. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-24 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ILQ
 
 | | 2.60A resolution structure of CT771 from Chlamydia trachomatis | | Descriptor: | CT771, GLYCEROL, SULFATE ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
4J22
 
 | | Tankyrase 2 in complex with 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide | | Descriptor: | 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4J34
 
 | | Crystal Structure of kynurenine 3-monooxygenase - truncated at position 394 plus HIS tag cleaved. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4IXU
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 11d: {(5R)-5-amino-5-carboxy-5-[(3-endo)-8-(3,4-dichlorobenzyl)-8-azabicyclo[3.2.1]oct-3-yl]pentyl}(trihydroxy)borate(1-) | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D, Beckett, P, Van Zandt, M.C, Ji, M.K, Ryder, T, Jagdmann, R, Andreoli, M, Olczak, J, Mazur, M, Czestkowski, W, Piotrowska, W, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of quaternary alpha-amino acid-based arginase inhibitors via the Ugi reaction.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
