6UTO
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
3BBV
 
 | |
8CG5
 
 | |
8CG4
 
 | |
8BY8
 
 | |
8CG6
 
 | |
6Q8X
 
 | | Respiratory complex I from Thermus thermophilus with bound Pyridaben. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-12-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6Q8W
 
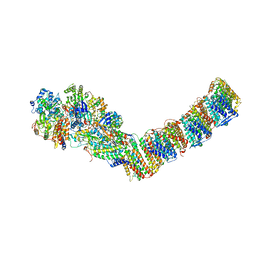 | | Respiratory complex I from Thermus thermophilus with bound Aureothin. | | Descriptor: | Aureothin, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-12-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6Q8O
 
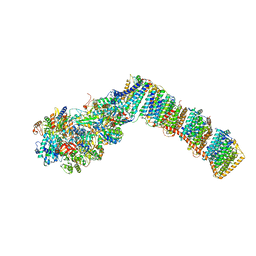 | | Respiratory complex I from Thermus thermophilus with bound Piericidin A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-12-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.605 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
5KKG
 
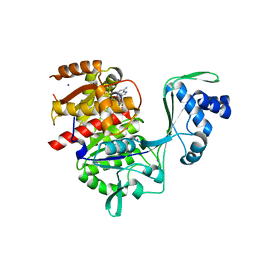 | | Crystal structure of E72A mutant of ancestral protein ancMT of ADP-dependent sugar kinases family | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, IODIDE ION, ... | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-06-21 | | Release date: | 2017-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
8UGC
 
 | |
6I0D
 
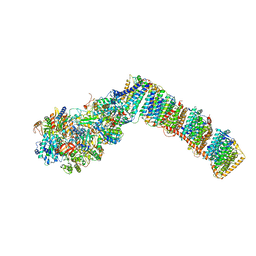 | | Respiratory complex I from Thermus thermophilus with bound Decyl-Ubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-10-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
5I3D
 
 | | Sulfolobus solfataricus beta-glycosidase - E387Y mutant | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ACETATE ION, Beta-galactosidase | | Authors: | Iglesias-Fernandez, J, Hancock, S.M, Lee, S.S, McAuley, K.E, Fordham-Skelton, A, Rovira, C, Davis, B.D. | | Deposit date: | 2016-02-10 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A front-face 'SNi synthase' engineered from a retaining 'double-SN2' hydrolase.
Nat. Chem. Biol., 13, 2017
|
|
6I1P
 
 | | Respiratory complex I from Thermus thermophilus with bound NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-10-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
5U8C
 
 | |
5TQ0
 
 | | Crystal structure of amino terminal domains of the NMDA receptor subunit GluN1 and GluN2A in the presence of EDTA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB, HEAVY CHAIN, ... | | Authors: | Romero-Hernandez, A, Simorowski, N, Karakas, E, Furukawa, H. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis for Subtype Specificity and High-Affinity Zinc Inhibition in the GluN1-GluN2A NMDA Receptor Amino-Terminal Domain.
Neuron, 92, 2016
|
|
5TSF
 
 | | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 7.0 spacegroup P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-N~4~,N~4~-bis[(pyridin-3-yl)methyl]pyrimidine-2,4-diamine, Soluble acetylcholine receptor | | Authors: | Camacho-Hernandez, G.A, Kaczanowska, K, Harel, M, Finn, M.G, Taylor, P.W. | | Deposit date: | 2016-10-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 7.0 spacegroup P212121
To Be Published
|
|
5TPZ
 
 | | Crystal structure of amino terminal domains of the NMDA receptor subunit GluN1 and GluN2B in apo closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 2B, ... | | Authors: | Romero-Hernandez, A, Simorwski, N, Karakas, E, Furukawa, H. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.095 Å) | | Cite: | Molecular Basis for Subtype Specificity and High-Affinity Zinc Inhibition in the GluN1-GluN2A NMDA Receptor Amino-Terminal Domain.
Neuron, 92, 2016
|
|
5TQ2
 
 | | Crystal structure of amino terminal domains of the NMDA receptor subunit GluN1 and GluN2A in complex with zinc at GluN1 and GluN2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB, HEAVY CHAIN, ... | | Authors: | Romero-Hernandez, A, Simorowski, N, Karakas, E, Furukawa, H. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.289 Å) | | Cite: | Molecular Basis for Subtype Specificity and High-Affinity Zinc Inhibition in the GluN1-GluN2A NMDA Receptor Amino-Terminal Domain.
Neuron, 92, 2016
|
|
5TPW
 
 | | Crystal structure of amino terminal domains of the NMDA receptor subunit GluN1 and GluN2A in complex with zinc at the GluN2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB, HEAVY CHAIN, ... | | Authors: | Romero-Hernandez, A, Simorowski, N, Karakas, E, Furukawa, H. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Molecular Basis for Subtype Specificity and High-Affinity Zinc Inhibition in the GluN1-GluN2A NMDA Receptor Amino-Terminal Domain.
Neuron, 92, 2016
|
|
5TVH
 
 | | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 8.0 spacegroup P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-N~4~,N~4~-bis[(pyridin-3-yl)methyl]pyrimidine-2,4-diamine, DIMETHYL SULFOXIDE, ... | | Authors: | Camacho-Hernandez, G.A, Kaczanowska, K, Harel, M, Finn, M.G, Taylor, P.W. | | Deposit date: | 2016-11-08 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 7.0 spacegroup P212121
To Be Published
|
|
8QCJ
 
 | |
8QCK
 
 | |
6Y11
 
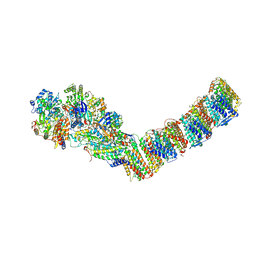 | | Respiratory complex I from Thermus thermophilus | | Descriptor: | DUF3197 domain-containing protein, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2020-02-10 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
7Q4Q
 
 | | Magacizumab Fab fragment in complex with human LRG1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRG1 epitope, Magacizumab heavy chain, ... | | Authors: | Gutierrez-Fernandez, J, Luecke, H. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of human LRG1 recognition by Magacizumab, a humanized monoclonal antibody with therapeutic potential.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
