6GA4
 
 | | Bacteriorhodopsin, 1 ps state, real-space refined against 15% extrapolated map | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAD
 
 | | BACTERIORHODOPSIN, 530 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
2ZE1
 
 | | X-ray structure of Bace-1 in complex with compound 6g | | Descriptor: | 3-bromo-N-[4-[1-(2-carbamimidamido-2-oxo-ethyl)-5-phenyl-pyrrol-2-yl]phenyl]benzamide, Beta-secretase 1 | | Authors: | Chopra, R, Olland, A. | | Deposit date: | 2007-12-05 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2G5W
 
 | | X-ray crystal structure of Arabidopsis thaliana 12-oxophytodienoate reductase isoform 3 (AtOPR3) in complex with 8-iso prostaglandin A1 and its cofactor, flavin mononucleotide. | | Descriptor: | (8S,12S)-15S-HYDROXY-9-OXOPROSTA-10Z,13E-DIEN-1-OIC ACID, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Han, B.W, Malone, T.E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Fox, B.G, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-23 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | Crystal structure of Arabidopsis thaliana 12-oxophytodienoate reductase isoform 3 in complex with 8-iso prostaglandin A(1).
Proteins, 79, 2011
|
|
1FMF
 
 | | REFINED SOLUTION STRUCTURE OF THE (13C,15N-LABELED) B12-BINDING SUBUNIT OF GLUTAMATE MUTASE FROM CLOSTRIDIUM TETANOMORPHUM | | Descriptor: | METHYLASPARTATE MUTASE S CHAIN | | Authors: | Hoffmann, B, Konrat, R, Tollinger, M, Huhta, M, Marsh, E.N.G, Kraeutler, B. | | Deposit date: | 2000-08-17 | | Release date: | 2002-02-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A protein pre-organized to trap the nucleotide moiety of coenzyme B(12): refined solution structure of the B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum.
Chembiochem, 2, 2001
|
|
3ENR
 
 | | ZINC-CALCIUM CONCANAVALIN A AT PH 6.15 | | Descriptor: | CALCIUM ION, Concanavalin-A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Wyns, L. | | Deposit date: | 1999-02-11 | | Release date: | 2000-09-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Zinc/calcium- and cadmium/cadmium-substituted concanavalin A: interplay of metal binding, pH and molecular packing.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3EQ4
 
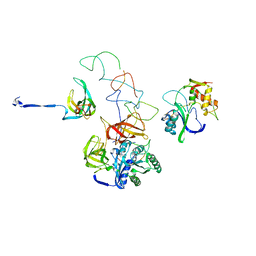 | | Model of tRNA(Leu)-EF-Tu in the ribosomal pre-accommodated state revealed by cryo-EM | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, Elongation factor Tu, ... | | Authors: | Frank, J, Li, W, Agirrezabala, X. | | Deposit date: | 2008-09-30 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM.
Embo J., 27, 2008
|
|
5AX3
 
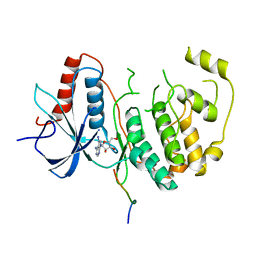 | | Crystal structure of ERK2 complexed with allosteric and ATP-competitive inhibitors. | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 1, allosteric and ATP-competitive inhibitor | | Authors: | Kinoshita, T, Sugiyama, H, Mori, Y, Takahashi, N, Tomonaga, A. | | Deposit date: | 2015-07-14 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Identification of allosteric ERK2 inhibitors through in silico biased screening and competitive binding assay
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6G7I
 
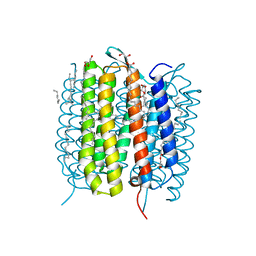 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 49-406 fs state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
1AMB
 
 | |
5VTM
 
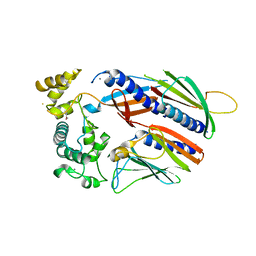 | |
1YFM
 
 | |
5B35
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T, Suzuki, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BOL
 
 | | DNA polymerase beta ternary complex with a templating 5ClC and incoming dGTP analog | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, DNA (5'-D(*CP*CP*GP*AP*CP*(CDO)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-05-27 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1AAP
 
 | | X-RAY CRYSTAL STRUCTURE OF THE PROTEASE INHIBITOR DOMAIN OF ALZHEIMER'S AMYLOID BETA-PROTEIN PRECURSOR | | Descriptor: | ALZHEIMER'S DISEASE AMYLOID A4 PROTEIN | | Authors: | Hynes, T.R, Randal, M, Kennedy, L.A, Eigenbrot, C, Kossiakoff, A.A. | | Deposit date: | 1990-09-14 | | Release date: | 1991-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of the protease inhibitor domain of Alzheimer's amyloid beta-protein precursor.
Biochemistry, 29, 1990
|
|
5BU8
 
 | |
5BUI
 
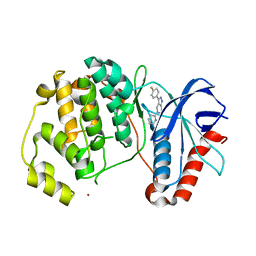 | | ERK2 complexed with 2-pyridiyl tetrahydroazaindazole | | Descriptor: | 3-(4-fluorophenyl)-5-(pyridin-2-yl)-4,5,6,7-tetrahydro-2H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase 1, NICKEL (II) ION | | Authors: | Bellamacina, C.R, Shu, W, Bussiere, D.E, Bagdanoff, J.T. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Ligand efficient tetrahydro-pyrazolopyridines as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3QTI
 
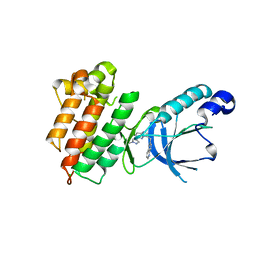 | | c-Met Kinase in Complex with NVP-BVU972 | | Descriptor: | 6-{[6-(1-methyl-1H-pyrazol-4-yl)imidazo[1,2-b]pyridazin-3-yl]methyl}quinoline, CHLORIDE ION, GLYCEROL, ... | | Authors: | Appleton, B.A. | | Deposit date: | 2011-02-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Drug Resistance Screen Using a Selective MET Inhibitor Reveals a Spectrum of Mutations That Partially Overlap with Activating Mutations Found in Cancer Patients.
Cancer Res., 71, 2011
|
|
5EB4
 
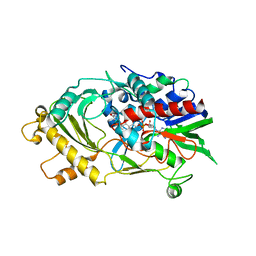 | | The crystal structure of almond HNL, PaHNL5 V317A, expressed in Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, K. | | Deposit date: | 2015-10-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of almond hydroxynitrile lyase isoenzyme 5 provide a rationale for the lack of oxidoreductase activity in flavin dependent HNLs.
J.Biotechnol., 235, 2016
|
|
3ND2
 
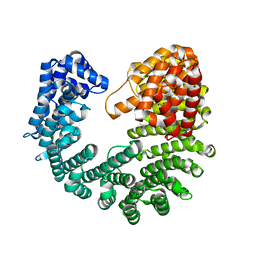 | | Structure of Yeast Importin-beta (Kap95p) | | Descriptor: | Importin subunit beta-1 | | Authors: | Forwood, J.K, Kobe, B. | | Deposit date: | 2010-06-06 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Quantitative Structural Analysis of Importin-beta Flexibility: Paradigm for Solenoid Protein Structures
Structure, 18, 2010
|
|
3EQ3
 
 | | Model of tRNA(Trp)-EF-Tu in the ribosomal pre-accommodated state revealed by cryo-EM | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, Elongation factor Tu, ... | | Authors: | Frank, J, Li, W, Agirrezabala, X. | | Deposit date: | 2008-09-30 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM.
Embo J., 27, 2008
|
|
1HYH
 
 | |
3CAO
 
 | | OXIDISED STRUCTURE OF THE ACIDIC CYTOCHROME C3 FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | ARSENIC, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Norager, S, Legrand, P, Pieulle, L, Hatchikian, C, Roth, M. | | Deposit date: | 1998-11-17 | | Release date: | 2000-07-23 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the oxidised and reduced acidic cytochrome c3from Desulfovibrio africanus.
J.Mol.Biol., 290, 1999
|
|
3GGX
 
 | | HIV Protease, pseudo-symmetric inhibitors | | Descriptor: | V-1 protease, methyl [(1S)-1-{[(1R,3S,4S)-4-{[(2S)-3,3-dimethyl-2-{3-[(6-methylpyridin-2-yl)methyl]-2-oxo-2,3-dihydro-1H-imidazol-1-yl}butanoyl]amino}-3-hydroxy-5-phenyl-1-(4-pyridin-2-ylbenzyl)pentyl]carbamoyl}-2,2-dimethylpropyl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
3GD4
 
 | |
