8OL1
 
 | |
5HIW
 
 | | Sorangium cellulosum So Ce56 cytochrome P450 260B1 | | Descriptor: | Cytochrome P450 CYP260B1, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Salamanca-Pinzon, S.G, Carius, Y, Khatri, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-01-12 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function analysis for the hydroxylation of Delta 4 C21-steroids by the myxobacterial CYP260B1.
Febs Lett., 590, 2016
|
|
8OQ4
 
 | |
6QNP
 
 | |
5HT0
 
 | | Crystal structure of an Antibiotic_NAT family aminoglycoside acetyltransferase HMB0038 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | Aminoglycoside acetyltransferase HMB0005, COENZYME A, SULFATE ION | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
2RIP
 
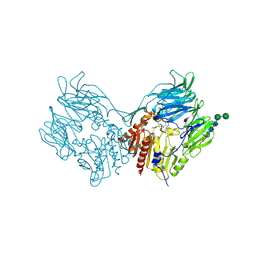 | | Structure of DPPIV in complex with an inhibitor | | Descriptor: | (3R,4R)-4-(pyrrolidin-1-ylcarbonyl)-1-(quinoxalin-2-ylcarbonyl)pyrrolidin-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Qiu, X. | | Deposit date: | 2007-10-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and synthesis of potent amido- and benzyl-substituted cis-3-amino-4-(2-cyanopyrrolidide)pyrrolidinyl DPP-IV inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5HX2
 
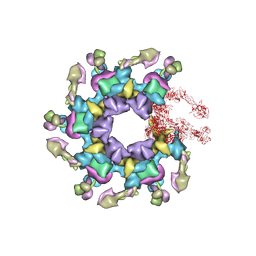 | | In vitro assembled star-shaped hubless T4 baseplate | | Descriptor: | Baseplate wedge protein gp10, Baseplate wedge protein gp53, Baseplate wedge protein gp6, ... | | Authors: | Yap, M.L, Klose, T, Fokine, A, Rossmann, M.G. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Role of bacteriophage T4 baseplate in regulating assembly and infection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6QP0
 
 | |
5HZP
 
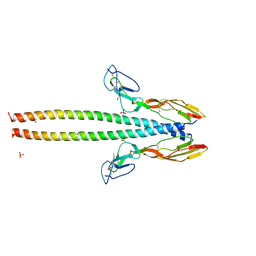 | | Structure of human C4b-binding protein alpha chain CCP domains 1 and 2 in complex with the hypervariable region of group A Streptococcus M49 protein. | | Descriptor: | C4b-binding protein alpha chain, M protein, serotype 49, ... | | Authors: | Buffalo, C.Z, Bahn-Suh, A.J, Ghosh, P. | | Deposit date: | 2016-02-02 | | Release date: | 2016-07-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein.
Nat Microbiol, 1, 2016
|
|
2RPS
 
 | | Solution structure of a novel insect chemokine isolated from integument | | Descriptor: | Chemokine | | Authors: | Kamiya, M, Nakatogawa, S, Oda, Y, Kamijima, T, Aizawa, T, Demura, M, Hayakawa, Y, Kawano, K. | | Deposit date: | 2008-07-28 | | Release date: | 2009-06-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A novel peptide mediates aggregation and migration of hemocytes from an insect
Curr.Biol., 19, 2009
|
|
2RPV
 
 | | Solution Structure of GB1 with LBT probe | | Descriptor: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | Authors: | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
5HTL
 
 | | Structure of MshE with cdg | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MSHA biogenesis protein MshE | | Authors: | Chin, K.H, Wang, Y.C. | | Deposit date: | 2016-01-27 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain.
Nat Commun, 7, 2016
|
|
5HHD
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D-Peptide RFX037.D, D-Vascular endothelial growth factor, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uppalapati, M, LEE, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
8OM6
 
 | |
5HX6
 
 | | Crystal structure of RIP1 kinase with a benzo[b][1,4]oxazepin-4-one | | Descriptor: | 5-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1,2-oxazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
2TMN
 
 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | Descriptor: | CALCIUM ION, N~2~-phosphono-L-leucinamide, Thermolysin, ... | | Authors: | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
6QXL
 
 | | Crystal Structure of Pyruvate Kinase II (PykA) from Pseudomonas aeruginosa in complex with sodium malonate, magnesium and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Abdelhamid, Y, Brear, P, Welch, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Evolutionary plasticity in the allosteric regulator-binding site of pyruvate kinase isoform PykA fromPseudomonas aeruginosa.
J.Biol.Chem., 294, 2019
|
|
5HIR
 
 | |
5HJF
 
 | |
8P1R
 
 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant. | | Descriptor: | 1,2-ETHANEDIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant, MAGNESIUM ION | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Exploring sequence, structure and function of microbial fucosidases from glycoside hydrolase GH29 family
To Be Published
|
|
2RDX
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Ozyurt, S, Gilmore, M, Lau, C, Maletic, M, Gheyi, T, Wasserman, S.R, Koss, J, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM.
To be Published
|
|
6QRH
 
 | |
5HOY
 
 | | X-ray crystallographic structure of an A-beta 17_36 beta-hairpin. X-ray diffractometer data set. (LVFFAEDCGSNKCAII(SAR)LMV). | | Descriptor: | Amyloid beta A4 protein, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Kreutzer, A.G, Spencer, R.K, Nowick, J.S. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | X-ray Crystallographic Structures of a Trimer, Dodecamer, and Annular Pore Formed by an A beta 17-36 beta-Hairpin.
J.Am.Chem.Soc., 138, 2016
|
|
6QSU
 
 | | Helicobacter pylori urease with BME bound in the active site | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2019-02-22 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
5HQO
 
 | | Crystal structure of IrCp*/I-Pd(allyl)-apo-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Maity, B, Fukumori, K, Abe, S, Ueno, T. | | Deposit date: | 2016-01-21 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Immobilization of two organometallic complexes into a single cage to construct protein-based microcompartments
Chem.Commun.(Camb.), 52, 2016
|
|
