4DKW
 
 | |
1P26
 
 | | Crystal structure of zinc(II)-d(GGCGCC)2 | | Descriptor: | 5'-D(*GP*GP*CP*GP*CP*C)-3', ZINC ION | | Authors: | Labiuk, S.L, Delbaere, L.T, Lee, J.S. | | Deposit date: | 2003-04-14 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Cobalt(II), nickel(II) and zinc(II) do not bind to intra-helical N(7)
guanine positions in the B-form crystal structure of d(GGCGCC)
J.Biol.Inorg.Chem., 8, 2003
|
|
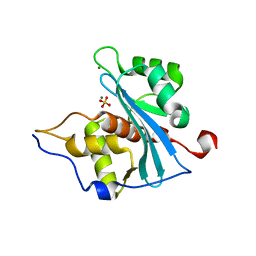
3NR1
 
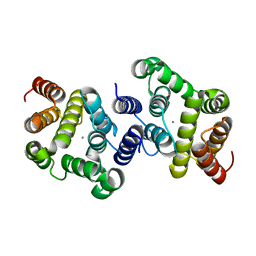 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | Descriptor: | HD domain-containing protein 3, MANGANESE (II) ION | | Authors: | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
5TBP
 
 | | Crystal Structure of RXR-alpha ligand binding domain complexed with synthetic modulator K8003 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Aleshin, A.E, Liddington, R.C, Su, Y, Zhang, X. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modulation of nongenomic activation of PI3K signalling by tetramerization of N-terminally-cleaved RXR alpha.
Nat Commun, 8, 2017
|
|
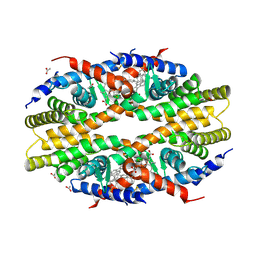
5W7W
 
 | | Crystal Structure of FHA domain of human APLF | | Descriptor: | Aprataxin and PNK-like factor, FORMIC ACID, SODIUM ION | | Authors: | Pedersen, L.C, Kim, K, London, R.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.348 Å) | | Cite: | Characterization of the APLF FHA-XRCC1 phosphopeptide interaction and its structural and functional implications.
Nucleic Acids Res., 45, 2017
|
|
7P2X
 
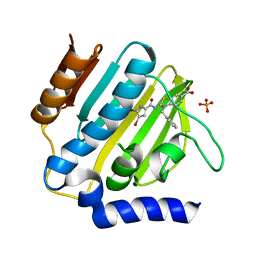 | | E.coli GyrB24 with inhibitor KOB20 (EBL2583) | | Descriptor: | (2Z)-2-[[4,5-bis(bromanyl)-1H-pyrrol-2-yl]carbonylimino]-3-(phenylmethyl)-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Benek, O, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | E.coli GyrB24 with inhibitor KOB20 (EBL2583)
TO BE PUBLISHED
|
|
7XYG
 
 | |
2HI3
 
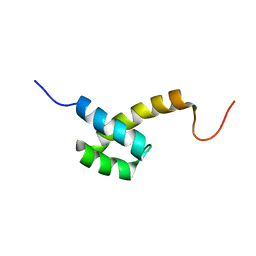 | | Solution structure of the homeodomain-only protein HOP | | Descriptor: | Homeodomain-only protein | | Authors: | Mackay, J.P, Kook, H, Epstein, J.A, Simpson, R.J, Yung, W.W. | | Deposit date: | 2006-06-28 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the structure and function of the transcriptional coregulator HOP
Biochemistry, 45, 2006
|
|
6ZQA
 
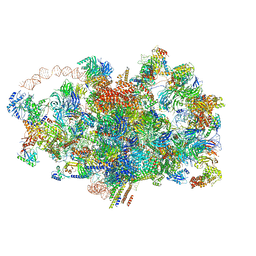 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state A (Poly-Ala) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQC
 
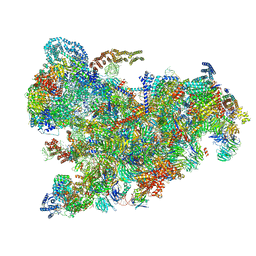 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Pre-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
4PSX
 
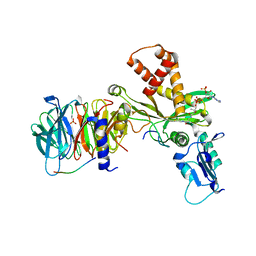 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
3TB6
 
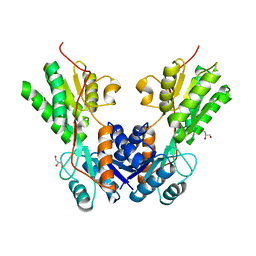 | |
3M66
 
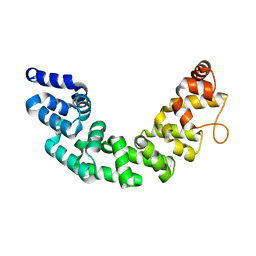 | | Crystal structure of human Mitochondrial Transcription Termination Factor 3 | | Descriptor: | mTERF domain-containing protein 1, mitochondrial | | Authors: | Spahr, H, Samuelsson, T, Hallberg, B.M, Gustafsson, C.M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain.
Biochem.Biophys.Res.Commun., 397, 2010
|
|
2HVD
 
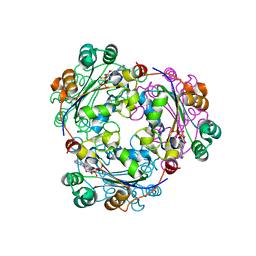 | | Human nucleoside diphosphate kinase A complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Giraud, M.-F, Georgescauld, F, Lascu, I, Dautant, A. | | Deposit date: | 2006-07-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of S120G Mutant and Wild Type of Human Nucleoside Diphosphate Kinase A in Complex with ADP
J.Bioenerg.Biomembr., 38, 2006
|
|
2HVE
 
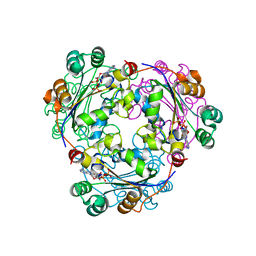 | | S120G mutant of human nucleoside diphosphate kinase A complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Giraud, M.-F, Georgescauld, F, Lascu, I, Dautant, A. | | Deposit date: | 2006-07-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal Structures of S120G Mutant and Wild Type of Human Nucleoside Diphosphate Kinase A in Complex with ADP
J.Bioenerg.Biomembr., 38, 2006
|
|
2HVQ
 
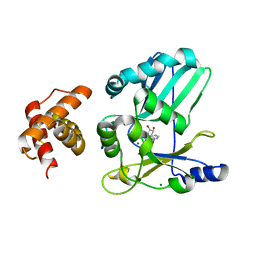 | | Structure of Adenylated full-length T4 RNA Ligase 2 | | Descriptor: | Hypothetical 37.6 kDa protein in Gp24-hoc intergenic region, MAGNESIUM ION | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
4AMS
 
 | |
2H4P
 
 | |
2HB5
 
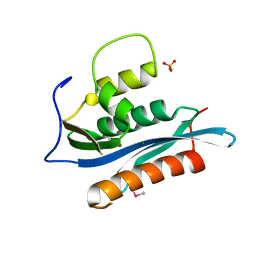 | | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain | | Descriptor: | MAGNESIUM ION, Reverse transcriptase/ribonuclease H, SULFATE ION | | Authors: | Lim, D, Gregorio, G.G, Bingman, C.A, Martinez-Hackert, E, Hendrickson, W.A, Goff, S.P. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain.
J.Virol., 80, 2006
|
|
1H6P
 
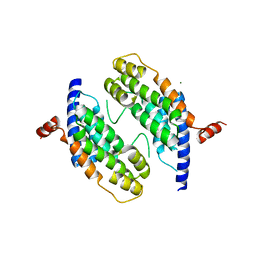 | | Dimeristion domain from human TRF2 | | Descriptor: | MAGNESIUM ION, TELOMERIC REPEAT BINDING FACTOR 2 | | Authors: | Chapman, L, Fairall, L, Rhodes, D. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Trfh Dimerization Domain of the Human Telomere Proteins Trf1 and Trf2
Mol.Cell, 8, 2001
|
|
4QNR
 
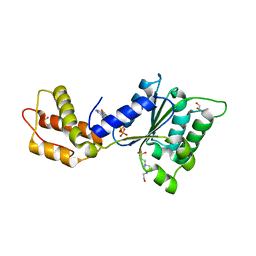 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ATP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QNM
 
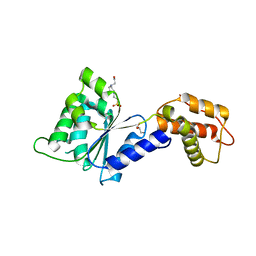 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4AMQ
 
 | |
4H75
 
 | | Crystal structure of human Spindlin1 in complex with a histone H3K4(me3) peptide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Histone H3, ... | | Authors: | Yang, N, Wang, W, Wang, Y, Wang, M, Zhao, Q, Rao, Z, Zhu, B, Xu, R.M. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ASL
 
 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ... | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
