8RFW
 
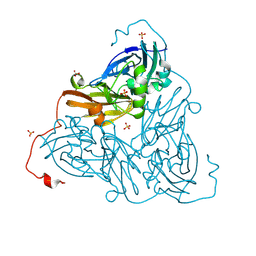 | | High pH (8.0) as-isolated MSOX movie series dataset 2 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [0.82 MGy] | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
5FLT
 
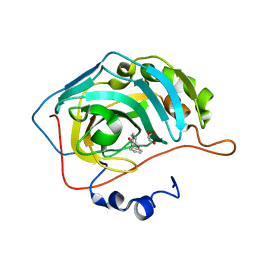 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 3-phenoxybenzoic acid, GLYCEROL, HUMAN CARBONIC ANHYDRASE 2, ... | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
7L2C
 
 | | Crystallographic structure of neutralizing antibody 2-51 in complex with SARS-CoV-2 spike N-terminal domain (NTD) | | Descriptor: | 2-51 heavy chain, 2-51 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cerutti, G, Reddem, E.R, Shapiro, L. | | Deposit date: | 2020-12-16 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
7PVW
 
 | |
6YIE
 
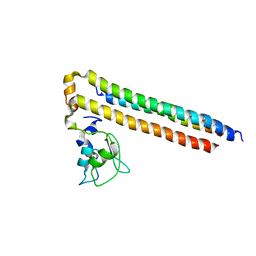 | | Structure of a Borealin-INCENP-Survivin complex | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, Inner centromere protein, ... | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
6FK3
 
 | | Structure and function of aldehyde dehydrogenase from Thermus thermophilus: An enzyme with an evolutionarily-distinct C-terminal arm (Recombinant full-length protein in complex with propanal) | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Aldehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hayes, K.A, Noor, M.R, Djeghader, A, Soulimane, T. | | Deposit date: | 2018-01-23 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The quaternary structure of Thermus thermophilus aldehyde dehydrogenase is stabilized by an evolutionary distinct C-terminal arm extension.
Sci Rep, 8, 2018
|
|
7ZPZ
 
 | | Crystal structure of Pizza6-TSR-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TSR-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
6JIS
 
 | | Crystal structure of the histidine racemase CntK in cobalt and nickel transporter system of staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CESIUM ION, CHLORIDE ION, ... | | Authors: | Luo, S, Ju, Y, Zhou, H. | | Deposit date: | 2019-02-23 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CntK, the cofactor-independent histidine racemase in staphylopine-mediated metal acquisition of Staphylococcus aureus.
Int.J.Biol.Macromol., 135, 2019
|
|
7ZNB
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 5.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PW0
 
 | |
7ZTI
 
 | |
6YIR
 
 | |
6JJ6
 
 | | BRD4 in complex with 500 | | Descriptor: | 1-methyl-6-(3-(4-methylpiperazine-1-carbonyl)benzyl)-1,2a1,5a,6-tetrahydro-2H-pyrido[3',2':6,7]azepino[4,3,2-cd]isoindol-2-one, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | BRD4 in complex with ZZM1
To Be Published
|
|
8QIY
 
 | |
6FL1
 
 | | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP(FOX)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B, Ober, M, Carell, T. | | Deposit date: | 2018-01-25 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA
To Be Published
|
|
7L3L
 
 | | Structure of TRAF5 and TRAF6 RING Hetero dimer | | Descriptor: | TNF receptor-associated factor 5, TNF receptor-associated factor 6, ZINC ION | | Authors: | Das, A, Middleton, A.J, Padala, P, Day, C.L. | | Deposit date: | 2020-12-17 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure and ubiquitin binding properties of TRAF RING heterodimers.
J.Mol.Biol., 2021
|
|
6BCT
 
 | | I-LtrI E184D bound to non-cognate C4 substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (26-MER), DNA (27-MER), ... | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
5UA5
 
 | |
5FYR
 
 | | Calcium-dependent phosphoinositol-specific phospholipase C from a Gram-negative bacterium, Pseudomonas sp, apo form, myoinositol complex | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Norgaard, A, Segura, D.R, Blicher, T.H, Wilson, K.S. | | Deposit date: | 2016-03-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a calcium-dependent phosphoinositide-specific phospholipase C from Pseudomonas sp. 62186, the first from a Gram-negative bacterium.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6PMZ
 
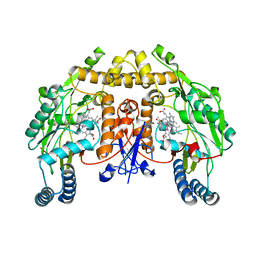 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(5-(Aminomethyl)pyridin-3-yl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[5-(aminomethyl)pyridin-3-yl]-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
7KV6
 
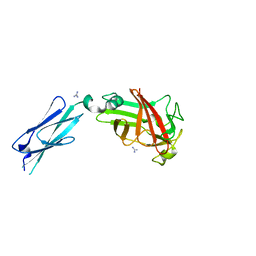 | |
7ZNA
 
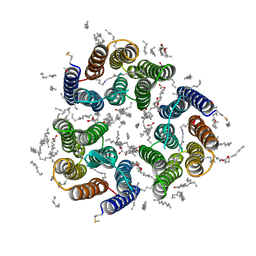 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 5.2 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5G11
 
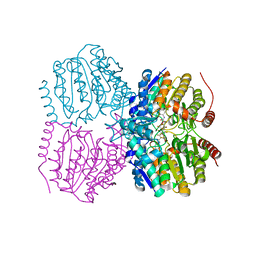 | | Pseudomonas aeruginosa HDAH bound to PFSAHA. | | Descriptor: | 2,2,3,3,4,4,5,5,6,6,7,7-dodecakis(fluoranyl)-~{N}-oxidanyl-~{N}'-phenyl-octanediamide, HDAH, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of a Histone Deacetylase Homologue from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
7KWB
 
 | |
7ZUA
 
 | |
