3V16
 
 | | An intramolecular pi-cation latch in phosphatidylinositol-specific phospholipase C from S.aureus controls substrate access to the active site | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, CHLORIDE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | Deposit date: | 2011-12-09 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
4OA7
 
 | | Crystal structure of Tankyrase1 in complex with IWR1 | | Descriptor: | 4-[(3aR,4R,7S,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Zhang, X, He, H. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
2RAH
 
 | | Human FDPS synthase in complex with novel inhibitor | | Descriptor: | Farnesyl pyrophosphate synthetase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Evdokimov, A.G, Barnett, B.L, Ebetino, F.H, Pokross, M. | | Deposit date: | 2007-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human FDPS synthase in complex with novel inhibitor
To be Published
|
|
4OAQ
 
 | | Crystal structure of the R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Aggrawal, N, Mandal, P.K, Gautham, N, Chadha, A. | | Deposit date: | 2014-01-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Insights into the stereoselectivity of R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330: Biochemical Characterization and Crystal structure studies
To be Published
|
|
4OLM
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-24 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
3V18
 
 | | Structure of the Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ISOPROPYL ALCOHOL, SULFATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | Deposit date: | 2011-12-09 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
2RFZ
 
 | | Crystal structure of cellobiohydrolase from Melanocarpus albomyces complexed with cellotriose | | Descriptor: | Cellulose 1,4-beta-cellobiosidase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
4OB3
 
 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila : A Reference Structure to Boronic Acid Inhibition of Nitrile Hydratase | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
3V14
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein complexed with Trehalose at 1.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein complexed with Trehalose at 1.70 A resolution
To be Published
|
|
2RGZ
 
 | | Ensemble refinement of the protein crystal structure of human heme oxygenase-2 C127A (HO-2) with bound heme | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bianchetti, C.M, Bingman, C.A, Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-10-05 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of Apo- and Heme-bound Crystal Structures of a Truncated Human Heme Oxygenase-2.
J.Biol.Chem., 282, 2007
|
|
3V1A
 
 | | Crystal structure of de novo designed MID1-apo1 | | Descriptor: | Computational design, MID1-apo1 | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
4G2P
 
 | | Crystal structure of peptidyl-prolyl cis-trans isomerase domain II of molecular chaperone SurA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | Chaperone SurA, GLYCEROL, SULFATE ION | | Authors: | Chang, C, Wu, R, Adkins, J.N, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-07-12 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of peptidyl-prolyl cis-trans isomerase domain II of molecular chaperone SurA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S
TO BE PUBLISHED
|
|
3V1Y
 
 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
4OM7
 
 | | Crystal structure of TIR domain of TLR6 | | Descriptor: | Toll-like receptor 6 | | Authors: | Park, H.H, Jang, T.H. | | Deposit date: | 2014-01-26 | | Release date: | 2014-08-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal Structure of TIR Domain of TLR6 Reveals Novel Dimeric Interface of TIR-TIR Interaction for Toll-Like Receptor Signaling Pathway.
J.Mol.Biol., 426, 2014
|
|
3V1E
 
 | | Crystal structure of de novo designed MID1-zinc H12E mutant | | Descriptor: | Computational design, MID1-zinc H12E mutant, ZINC ION | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.073 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
4OMA
 
 | | The crystal structure of methionine gamma-lyase from Citrobacter freundii in complex with L-cycloserine pyridoxal-5'-phosphate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2014-01-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pre-steady-state Kinetic and Structural Analysis of Interaction of Methionine gamma-Lyase from Citrobacter freundii with Inhibitors.
J.Biol.Chem., 290, 2015
|
|
4OC0
 
 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CCIBzL, a urea-based inhibitor N~2~-[(1-carboxycyclopropyl)carbamoyl]-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3V1O
 
 | |
4OCQ
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3V2U
 
 | | Crystal structure of the yeast GAL regulon complex of the repressor, Gal80p, and the transducer, Gal3p, with galactose and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Galactose/lactose metabolism regulatory protein GAL80, ... | | Authors: | Lavy, T, Kumar, P.R, He, H, Joshua-Tor, L. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation.
Genes Dev., 26, 2012
|
|
3V1V
 
 | |
4OD6
 
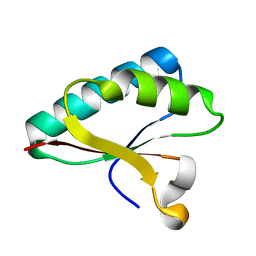 | | Structure of Smr domain of MutS2 from Deinococcus radiodurans, Mn2+ soaked | | Descriptor: | Endonuclease MutS2 | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
3V2G
 
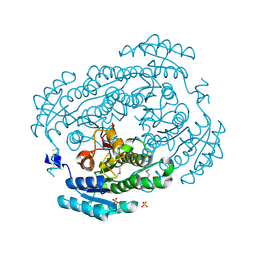 | | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, SULFATE ION | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-01-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021
To be Published
|
|
4OMW
 
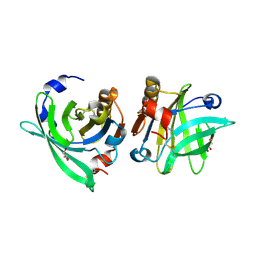 | | Crystal structure of goat beta-lactoglobulin (orthorhombic form) | | Descriptor: | Beta-lactoglobulin, GLYCEROL, SULFATE ION, ... | | Authors: | Loch, J.I, Swiatek, S, Czub, M, Ludwikowska, M, Lewinski, K. | | Deposit date: | 2014-01-27 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational variability of goat beta-lactoglobulin: Crystallographic and thermodynamic studies.
Int.J.Biol.Macromol., 72C, 2014
|
|
4G4X
 
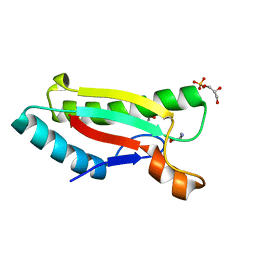 | | Crystal structure of peptidoglycan-associated lipoprotein from Acinetobacter baumannii | | Descriptor: | D-ALANINE, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
To be Published
|
|
