4CCN
 
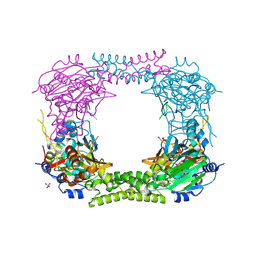 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 L299C/C300S) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-2) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
3CAS
 
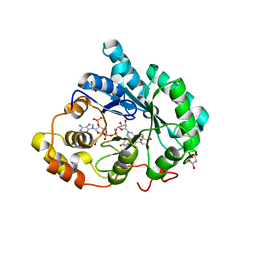 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADP+ and 4-androstenedione | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, 4-ANDROSTENE-3-17-DIONE, ... | | Authors: | Faucher, F, Cantin, L, Breton, R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Delta4-3-Ketosteroid 5beta-Reductase (AKR1D1) Reveal the Presence of an Alternative Binding Site Responsible for Substrate Inhibition (dagger) (,) (double dagger).
Biochemistry, 47, 2008
|
|
1L8A
 
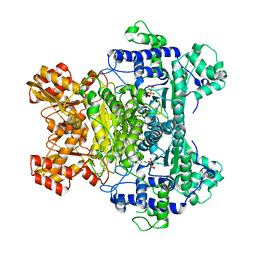 | | E. COLI PYRUVATE DEHYDROGENASE | | Descriptor: | MAGNESIUM ION, Pyruvate dehydrogenase E1 component, THIAMINE DIPHOSPHATE | | Authors: | Furey, W, Arjunan, P. | | Deposit date: | 2002-03-19 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 A resolution.
Biochemistry, 41, 2002
|
|
4NJO
 
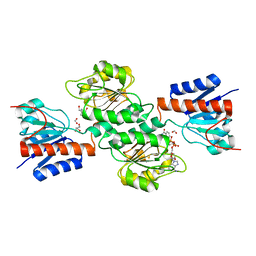 | |
5IVJ
 
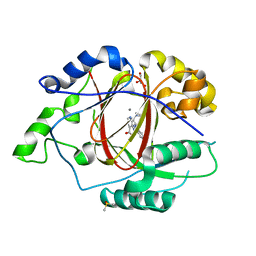 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N11 [3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid] | | Descriptor: | 3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IWE
 
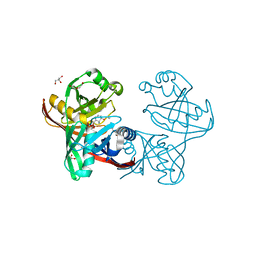 | | E45Q mutant of phenazine biosynthesis protein PhzF in complex with (5R,6R)-6-azaniumyl-5-ethoxycyclohexa-1,3-diene-1-carboxylate | | Descriptor: | (5R,6R)-6-azaniumyl-5-ethoxycyclohexa-1,3-diene-1-carboxylate, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Diederich, C, Blankenfeldt, W. | | Deposit date: | 2016-03-22 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mechanisms and Specificity of Phenazine Biosynthesis Protein PhzF.
Sci Rep, 7, 2017
|
|
7CZI
 
 | |
1QU9
 
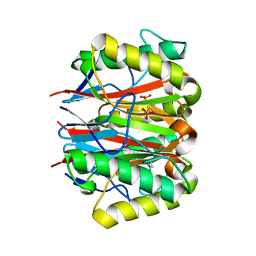 | | 1.2 A CRYSTAL STRUCTURE OF YJGF GENE PRODUCT FROM E. COLI | | Descriptor: | YJGF PROTEIN | | Authors: | Volz, K. | | Deposit date: | 1999-07-07 | | Release date: | 1999-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A test case for structure-based functional assignment: the 1.2 A crystal structure of the yjgF gene product from Escherichia coli
Protein Sci., 8, 1999
|
|
3ZQA
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE MUTANT C286A FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, BETAINE ALDEHYDE DEHYDROGENASE, ... | | Authors: | Diaz-Sanchez, A.G, Gonzalez-Segura, L, Rudino-Pinera, E, Lira-Rocha, A, Torres-Larios, A, Munoz-Clares, R.A. | | Deposit date: | 2011-06-08 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel Nadph-Cysteine Covalent Adduct Found in the Active Site of an Aldehyde Dehydrogenase.
Biochem.J., 439, 2011
|
|
6LTR
 
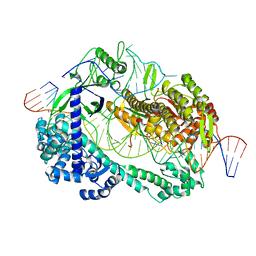 | | Crystal structure of Cas12i2 ternary complex with single Mg2+ bound in catalytic pocket | | Descriptor: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
5CVE
 
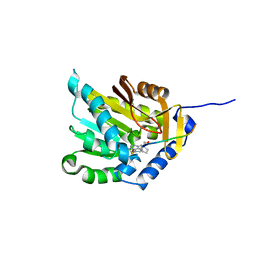 | |
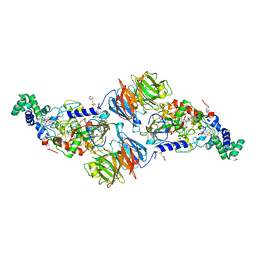
4O1Q
 
 | |
4ILK
 
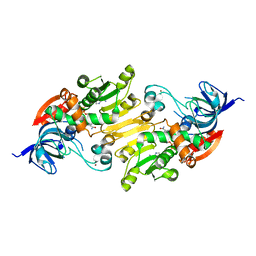 | | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MANGANESE (II) ION, Starvation sensing protein rspB, ... | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-12-31 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH
To be Published
|
|
2VHZ
 
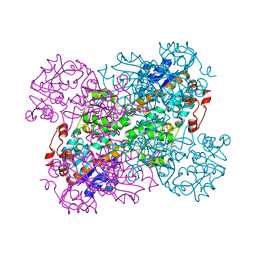 | |
4R41
 
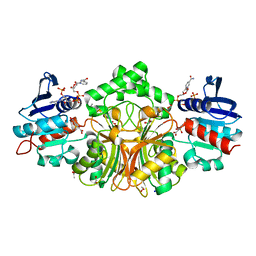 | | Complex Crystal structure of 4-nitro-2-phosphono-benzoic acid with sp-Aspartate-Semialdehyde Dehydrogenase and Nicotinamide-dinucleotide | | Descriptor: | 1,2-ETHANEDIOL, 4-nitro-2-phosphonobenzoic acid, ACETATE ION, ... | | Authors: | Pavlovsky, A.G, Viola, R.E. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3T0G
 
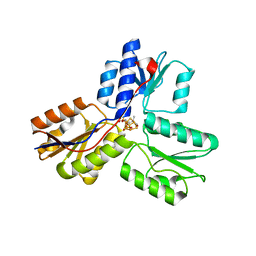 | | IspH:HMBPP (substrate) structure of the T167C mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
5GGF
 
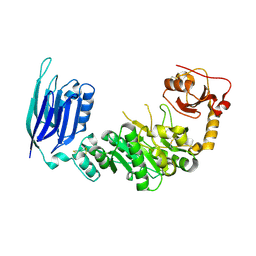 | |
2VHW
 
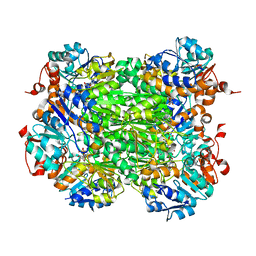 | |
2VAO
 
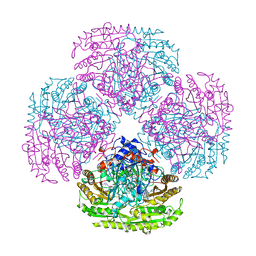 | |
4R51
 
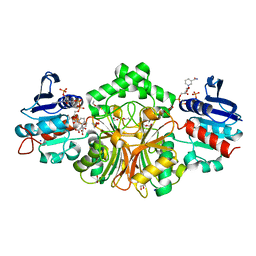 | | Crystal complex structure of sp-Aspartate-Semialdehyde-Dehydrogenase with Nicotinamide Adenine dinucleotide phosphate and phthalic acid | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Viola, R.E. | | Deposit date: | 2014-08-20 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3IBO
 
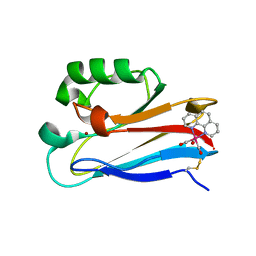 | | Pseudomonas aeruginosa E2Q/H83Q/T126H-azurin RE(PHEN)(CO)3 | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Gradinaru, C, Crane, B.R, Sudhamsu, J. | | Deposit date: | 2009-07-16 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Relaxation dynamics of Pseudomonas aeruginosa Re(I)(CO)3(alpha-diimine)(HisX)+ (X = 83, 107, 109, 124, 126)Cu(II) azurins.
J.Am.Chem.Soc., 131, 2009
|
|
6M5X
 
 | | A fungal glyceraldehyde-3-phosphate dehydrogenase with self-resistance to inhibitor heptelidic acid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05990934 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
6LY4
 
 | | The crystal structure of the BM3 mutant LG-23 in complex with testosterone | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, ... | | Authors: | Peng, Y, Chen, J, Zhou, J, Li, A, ReetZ, M.T. | | Deposit date: | 2020-02-13 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Regio- and Stereoselective Steroid Hydroxylation at C7 by Cytochrome P450 Monooxygenase Mutants.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4BL5
 
 | | Crystal structure of human GDP-L-fucose synthase with bound NADP and product GDP-L-fucose | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-FUCOSE SYNTHASE, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, ... | | Authors: | Vollmar, M, Shafqat, N, Rojkova, A, Krojer, T, Bradley, A, Raynor, J.W, Kavanagh, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U, Yue, W.W. | | Deposit date: | 2013-05-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human Gdp-L-Fucose Synthase with Bound Nadp and Product Gdp-L-Fucose
To be Published
|
|
5ZE0
 
 | | Hairpin Forming Complex, RAG1/2-Nicked(with Dideoxy) 12RSS/23RSS complex in Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (39-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
