8PO3
 
 | |
5WOP
 
 | | High Resolution Structure of Mutant CA09-PB2cap | | Descriptor: | GLYCEROL, Polymerase PB2 | | Authors: | Constantinides, A.E, Gumpper, R.H, Severin, C, Luo, M. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution structure of the Influenza A virus PB2cap binding domain illuminates the changes induced by ligand binding.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5WI9
 
 | | Crystal structure of KL with an agonist Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 39F7 Fab heavy chain, ... | | Authors: | Johnstone, S, Min, X, Wang, Z. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Agonistic beta-Klotho antibody mimics fibroblast growth factor 21 (FGF21) functions.
J. Biol. Chem., 293, 2018
|
|
8DGP
 
 | | 14-3-3 epsilon bound to phosphorylated PEAK3 (pS69) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Phosphorylated PEAK3 (pS69) peptide, ... | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
5LK3
 
 | |
8PO1
 
 | |
7XIU
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
5LKV
 
 | |
8D7I
 
 | |
7XIT
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
8PI3
 
 | | Cathepsin S Y132D mutant in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 20, 2024
|
|
7XJ7
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
5WK7
 
 | | P450cam mutant R186A | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Batabyal, D, Poulos, T.L. | | Deposit date: | 2017-07-24 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Effect of Redox Partner Binding on Cytochrome P450 Conformational Dynamics.
J. Am. Chem. Soc., 139, 2017
|
|
7XJ5
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
8PWX
 
 | |
5EJE
 
 | | Crystal structure of E. coli Adenylate kinase G56C/T163C double mutant in complex with Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, COBALT (II) ION | | Authors: | Sauer, U.H, Kovermann, M, Grundstrom, C, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2015-11-01 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for ligand binding to an enzyme by a conformational selection pathway.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7XJ4
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, S-[5-[(E)-2-phenylethenyl]-1,3,4-oxadiazol-2-yl] 5-nitrothiophene-2-carbothioate, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
5WKI
 
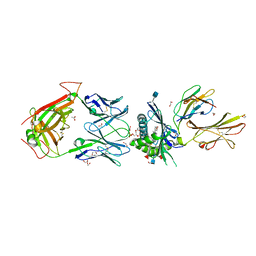 | | Crystal structure of PG90 TCR-CD1b-PG complex | | Descriptor: | (19S,22R,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shahine, A, Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A molecular basis of human T cell receptor autoreactivity toward self-phospholipids.
Sci Immunol, 2, 2017
|
|
5M19
 
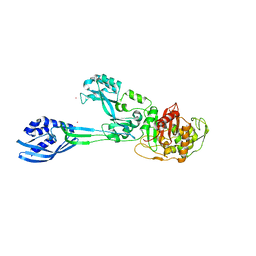 | | Crystal structure of PBP2a from MRSA in the presence of Oxacillin ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
7XIS
 
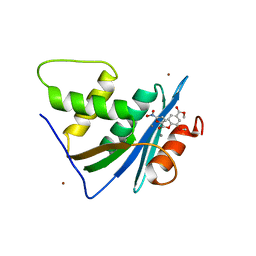 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | (2-methoxy-4-methoxycarbonyl-phenyl) 5-nitrofuran-2-carboxylate, MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
8PAA
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2552 | | Descriptor: | 7-[(1~{S})-1-[4-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]phenyl]carbonyloxyethyl]-3-[6-(morpholin-4-ylmethyl)pyridin-3-yl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2552
To Be Published
|
|
8PAN
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2653 | | Descriptor: | 7-[(1~{S})-1-[4-(aminomethyl)phenoxy]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2653
To Be Published
|
|
5EJM
 
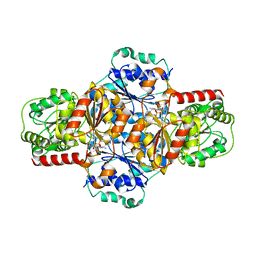 | | ThDP-Mn2+ complex of R413A variant of EcMenD soaked with 2-ketoglutarate for 35 min | | Descriptor: | (4~{R})-4-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3-ium-2-yl]-4-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Two active site arginines are critical determinants of substrate binding and catalysis in MenD, a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 2018
|
|
5WZT
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 14 | | Descriptor: | 2-[1-[(3-bromophenyl)methyl]-2-methyl-3-oxamoyl-indol-4-yl]oxyethanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5M3E
 
 | | Macrodomain of Thermus aquaticus DarG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Appr-1-p processing domain protein, CHLORIDE ION | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
