2MW9
 
 | |
2MWE
 
 | | NMR structure of FBP28 WW2 mutant Y438R, L453A DNDC | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MUM
 
 | | Solution structure of the PHD domain of Yeast YNG2 | | Descriptor: | Chromatin modification-related protein YNG2, ZINC ION | | Authors: | Taeb, S, Kaustov, L, Lemak, A, Farhadi, S, Sheng, Y. | | Deposit date: | 2014-09-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain of Yeast YNG2
To be Published
|
|
2N8S
 
 | |
2N8T
 
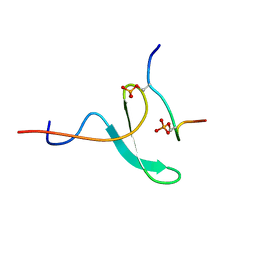 | |
2NNV
 
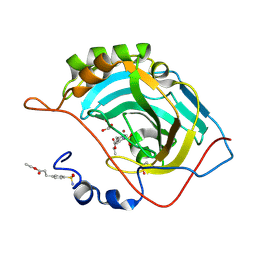 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase 2, ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2N0H
 
 | | NMR structure of Neuromedin C in presence of SDS micelles | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2QWK
 
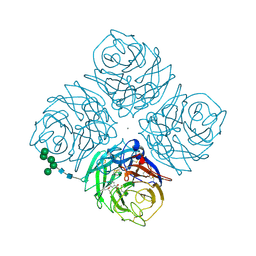 | |
2QB3
 
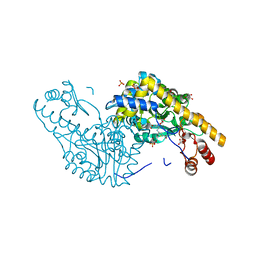 | | Structural Studies Reveal the Inactivation of E. coli L-Aspartate Aminotransferase by (s)-4,5-dihydro-2-thiophenecarboxylic acid (SADTA) via Two Mechanisms (at pH 7.5) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
2QCB
 
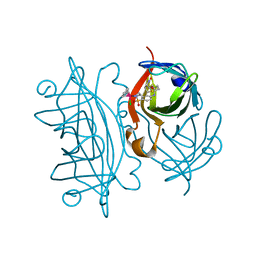 | | T7-tagged full-length streptavidin complexed with ruthenium ligand | | Descriptor: | N-(4-{[(2-AMINOETHYL)AMINO]SULFONYL}PHENYL)-5-[(3AS,4S,6AR)-2-OXOHEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL]PENTANAMIDE-(1,2,3,4,5,6-ETA)-BENZENE-CHLORO-RUTHENIUM(III), Streptavidin | | Authors: | Le Trong, I, Creus, M, Pordea, A, Ward, T.R, Stenkamp, R.E. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and designed evolution of an artificial transfer hydrogenase
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
2QJJ
 
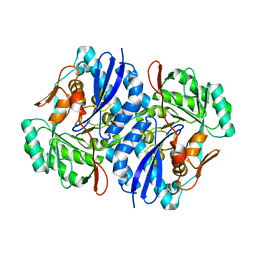 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
2QP3
 
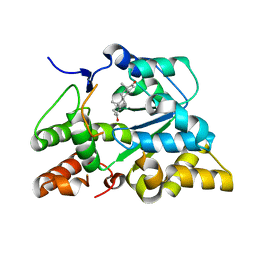 | |
2QWH
 
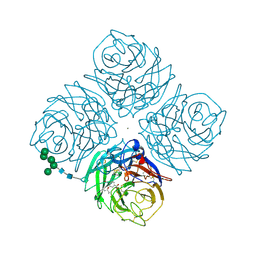 | |
2QWA
 
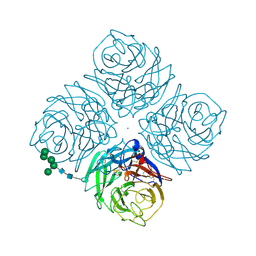 | |
2QWJ
 
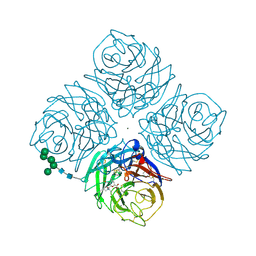 | |
2QZ3
 
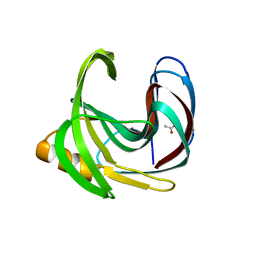 | | Crystal structure of a glycoside hydrolase family 11 xylanase from Bacillus subtilis in complex with xylotetraose | | Descriptor: | ACETIC ACID, Endo-1,4-beta-xylanase A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
2MDI
 
 | | Solution structure of the PP2WW mutant (KPP2WW) of HYPB | | Descriptor: | Histone-lysine N-methyltransferase SETD2 | | Authors: | Gao, Y.G, Hu, H.Y. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin.
Structure, 22, 2014
|
|
2MEJ
 
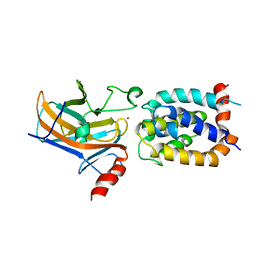 | |
2M3O
 
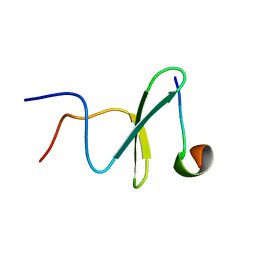 | | Structure and dynamics of a human Nedd4 WW domain-ENaC complex | | Descriptor: | Amiloride-sensitive sodium channel subunit alpha, E3 ubiquitin-protein ligase NEDD4 | | Authors: | Bobby, R, Medini, K, Neudecker, P, Lee, V, MacDonald, F.J, Brimble, M.A, Lott, J, Dingley, A.J. | | Deposit date: | 2013-01-23 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of human Nedd4-1 WW3 in complex with the alpha ENaC PY motif.
Biochim.Biophys.Acta, 1834, 2013
|
|
2M6Z
 
 | |
2QBT
 
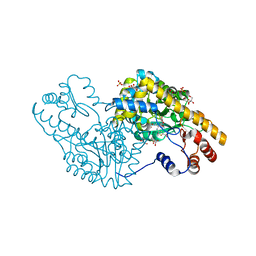 | | Structural Studies Reveal The Inactivation of E. coli L-aspartate aminotransferase by (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via Two Mechanisms (at pH 8.0) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inactivation of Escherichia coli l-Aspartate Aminotransferase by (S)-4-Amino-4,5-dihydro-2-thiophenecarboxylic Acid Reveals "A Tale of Two Mechanisms".
Biochemistry, 46, 2007
|
|
2QLX
 
 | |
2QRY
 
 | | Periplasmic thiamin binding protein | | Descriptor: | THIAMIN PHOSPHATE, Thiamine-binding periplasmic protein | | Authors: | Ealick, S.E, Soriano, E.V. | | Deposit date: | 2007-07-30 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Similarities between Thiamin-Binding Protein and Thiaminase-I Suggest a Common Ancestor
Biochemistry, 47, 2008
|
|
2QZY
 
 | | The structure of chicken mitochondrial PEPCK in complex with PEP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Holyoak, T, Sullivan, S.M, Nowak, T. | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Mechanism of PEPCK Catalysis
Biochemistry, 45, 2006
|
|
2QA3
 
 | | Structural Studies Reveal the Inactivation of E. coli L-aspartate aminotransferase by (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via two mechanisms (at pH6.5) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
