6FKT
 
 | |
6W4P
 
 | | CaMKII alpha-30 Cryo-EM reconstruction - Class B | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Chao, L.H, Stratton, M.M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub-dependent regulation.
Sci.Signal., 13, 2020
|
|
8IEJ
 
 | | RNF20-RNF40/hRad6A-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Ai, H, Deng, Z, Sun, M, Du, Y, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
6SU0
 
 | | Crystal structure of dimethylated RSLex in complex with cucurbit[7]uril | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, cucurbit[7]uril, ... | | Authors: | Guagnini, F, Engilberge, S, Crowley, P.B. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineered assembly of a protein-cucurbituril biohybrid.
Chem.Commun.(Camb.), 56, 2020
|
|
7RIN
 
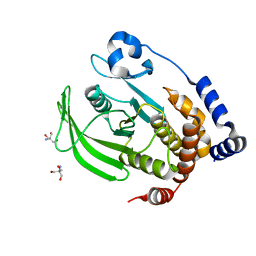 | |
6VVO
 
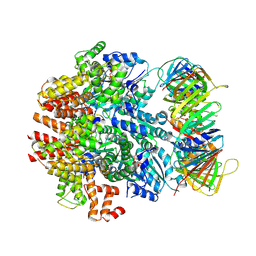 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
5UD2
 
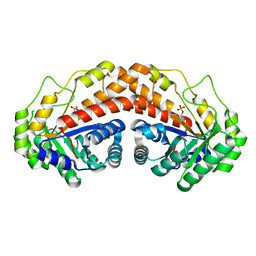 | | Class II fructose-1,6-bisphosphate aldolase H180Q variant of Helicobacter pylori with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
6J9V
 
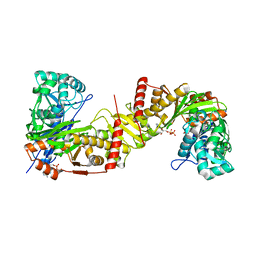 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reaction mechanism of the reverse reaction of African human trypanosomes glycerol kinase.
To Be Published
|
|
5UFX
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with OP1074 | | Descriptor: | (2S)-3-(4-hydroxyphenyl)-4-methyl-2-(4-{2-[(3R)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-2H-1-benzopyran-7-ol, Estrogen receptor | | Authors: | Fanning, S.W, Hodges-Gallagher, L, Myles, D.C, Sun, R, Fowler, C.E, Green, B.D, Harmon, C.L, Greene, G.L, Kushner, P.J. | | Deposit date: | 2017-01-06 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5503 Å) | | Cite: | Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity.
Nat Commun, 9, 2018
|
|
8T3N
 
 | | Solution NMR structure of synthetic peptide AMPCry10Aa_5 rational designed from Cry10Aa bacterial protein | | Descriptor: | Pesticidal crystal protein Cry10Aa peptide | | Authors: | Barra, J.B, Freitas, C.D.P, Rios, T.B, Maximiano, M.R, Fernandes, F.C, Amorim, G.C, Porto, W.F, Grossi-de-Sa, M.F, Franco, O.F, Liao, L.M. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | SOLUTION NMR | | Cite: | Anti-Staphy Peptides Rationally Designed from Cry10Aa Bacterial Protein.
Acs Omega, 9, 2024
|
|
6BIA
 
 | | Crystal structure of Ps i-CgsB | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-11-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
8TSO
 
 | | KDL bound, nucleotide-free MsbA in open, outward-facing conformation | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-binding transport protein MsbA, PENTAETHYLENE GLYCOL MONODECYL ETHER | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Native mass spectrometry captures snapshots of the MsbA transport cycle
To Be Published
|
|
6BTE
 
 | | DNA Polymerase Beta I260Q Binary Complex | | Descriptor: | CHLORIDE ION, DNA Downstream Strand, DNA Primer Strand, ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2017-12-06 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | I260Q DNA polymerase beta highlights precatalytic conformational rearrangements critical for fidelity.
Nucleic Acids Res., 46, 2018
|
|
4R4J
 
 | | Crystal structure of complex sp_ASADH with 3-carboxypropyl-phthalic acid and Nicotinamide Adenine dinucleotide phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-carboxypropyl)benzene-1,2-dicarboxylic acid, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Thangavelu, B, Bhansali, P, Viola, R.E. | | Deposit date: | 2014-08-19 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7RBQ
 
 | | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9, ... | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor
To Be Published
|
|
6W3U
 
 | | APE1 exonuclease substrate complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6BU6
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a bis-difluorophenol-aminopyridine inhibitor | | Descriptor: | 4,4'-(2-aminopyridine-3,5-diyl)bis(2,6-difluorophenol), CHLORIDE ION, GLYCEROL, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Santiago, A.S, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-08 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a bis-difluorophenol-aminopyridine inhibitor
To Be Published
|
|
6JIF
 
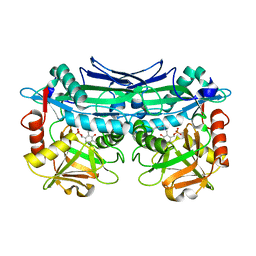 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-01 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
6W72
 
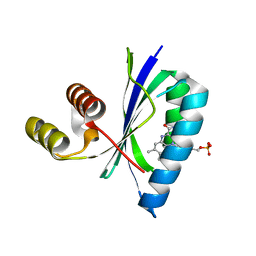 | | BlsA photo-activated state | | Descriptor: | BLUF domain-containing protein, FLAVIN MONONUCLEOTIDE | | Authors: | Chitrakar, I, French, J.B. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for the Regulation of Biofilm Formation and Iron Uptake in A. baumannii by the Blue-Light-Using Photoreceptor, BlsA.
Acs Infect Dis., 6, 2020
|
|
8T3H
 
 | | Solution NMR structure alpha-helix 3 of Cry10Aa protein | | Descriptor: | Pesticidal crystal protein Cry10Aa peptide | | Authors: | Barra, J.B, Freitas, C.D.P, Rios, T.B, Maximiano, M.R, Fernandes, F.C, Amorim, G.C, Porto, W.F, Grossi-de-Sa, M.F, Franco, O.F, Liao, L.M. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Alpha-helix 3 of Cry10Aa protein
To Be Published
|
|
8CE9
 
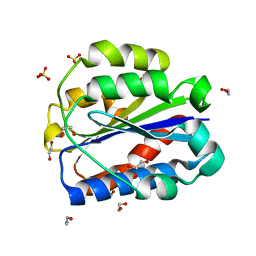 | | Crystal structure of human Cd11b I domain in C121 space group | | Descriptor: | 1,2-ETHANEDIOL, Integrin alpha-M, SULFATE ION | | Authors: | Koekemoer, L, Williams, E, Ni, X, Gao, Q, Coker, J, MacLean, E.M, Wright, N.D, Marsden, B, Von Delft, F, Schwenzer, A, Midwood, K. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of human Cd11b I domain in C121 space group
To Be Published
|
|
8ACM
 
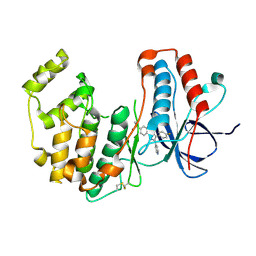 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACO
 
 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
5FDR
 
 | | Mcl-1 complexed with small molecule inhibitor | | Descriptor: | 5-[[6-chloranyl-3-[3-(4-chloranyl-3,5-dimethyl-phenoxy)propyl]-7-(3,5-dimethyl-1~{H}-pyrazol-4-yl)-1~{H}-indol-2-yl]carbonylsulfamoyl]furan-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods.
J.Med.Chem., 59, 2016
|
|
