6JFQ
 
 | |
5MXQ
 
 | | Crystal Structure of the Acquired VIM-2 Metallo-beta-Lactamase in Complex with ANT-90 Inhibitor | | Descriptor: | 3-(phenylsulfonylamino)pyridine-2-carboxylic acid, ACETATE ION, Beta-lactamase VIM-2, ... | | Authors: | Docquier, J.D, De Luca, F, Benvenuti, M, Di Pisa, F, Pozzi, C, Mangani, S. | | Deposit date: | 2017-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAR Studies Leading to the Identification of a Novel Series of Metallo-beta-lactamase Inhibitors for the Treatment of Carbapenem-Resistant Enterobacteriaceae Infections That Display Efficacy in an Animal Infection Model.
Acs Infect Dis., 5, 2019
|
|
6JMS
 
 | | CJP38, a beta-1,3-glucanase and allergen of Cryptomeria japonica pollen | | Descriptor: | 1,2-ETHANEDIOL, Pollen allergen CJP38 | | Authors: | Takashima, T, Numata, T, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CJP38, a beta-1,3-glucanase and allergen of Cryptomeria japonica pollen
To Be Published
|
|
5U7V
 
 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Trifolium repens in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Apyrase | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
5KBG
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH OCRESOL | | Descriptor: | MopR, ZINC ION, o-cresol | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
6JM5
 
 | | Crystal structure of TBC1D23 C terminal domain | | Descriptor: | SODIUM ION, TBC1 domain family member 23 | | Authors: | Sun, Q, Huang, W. | | Deposit date: | 2019-03-07 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5MSE
 
 | | GFP nuclear transport receptor mimic 3B8 | | Descriptor: | Green fluorescent protein, IMIDAZOLE, SODIUM ION | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2017-01-04 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Surface Properties Determining Passage Rates of Proteins through Nuclear Pores.
Cell, 174, 2018
|
|
6JNI
 
 | |
5U9A
 
 | |
1F74
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II COMPLEXED WITH 4-DEOXY-SIALIC ACID | | Descriptor: | 6,7,8,9-TETRAHYDROXY-5-METHYLCARBOXAMIDO-2-OXONONANOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-26 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
6JOH
 
 | |
5KDU
 
 | | ZmpB metallopeptidase in complex with a2,6-Sialyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, SERINE, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MT9
 
 | | Human insulin in complex with serotonin and arginine | | Descriptor: | ARGININE, CHLORIDE ION, Insulin, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2017-01-07 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
6JOW
 
 | |
5MU0
 
 | | ACC1 Fab fragment in complex with citrullinated C1 epitope of CII (IA03) | | Descriptor: | IA03 peptide containing the citrullinated C1 epitope of collagen type II,Collagen alpha-1(II) chain,IA03 peptide containing the citrullinated C1 epitope of collagen type II, heavy chain of ACC1 antibody Fab fragment, light chain of ACC1 antibody Fab fragment | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-11 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5JV4
 
 | | Structure of F420 binding protein, MSMEG_6526, from Mycobacterium smegmatis with F420 bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COENZYME F420, ... | | Authors: | Lee, B.M, Carr, P.D, Jackson, C.J. | | Deposit date: | 2016-05-10 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of F420 binding protein, MSMEG_6526, from Mycobacterium smegmatis with F420 bound
To Be Published
|
|
5MVV
 
 | | Crystal structure of Plasmodium falciparum actin I- gelsolin segment 1 -CdATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CADMIUM ION, ... | | Authors: | Panneerselvam, S, Kumpula, E.-P, Kursula, I, Burkhardt, A, Meents, A. | | Deposit date: | 2017-01-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid cadmium SAD phasing at the standard wavelength (1 angstrom ).
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1F7B
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II IN COMPLEX WITH 4-OXO-SIALIC ACID | | Descriptor: | 4,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDONONANOIC ACID, 6,7,8,9-TETRAHYDROXY-5-METHYLCARBOXAMIDO-4-OXONONANOIC ACID, CHLORIDE ION, ... | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-26 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
6JRC
 
 | | ZHD complex with hydrolyzed alpha-ZOL | | Descriptor: | 2-[(~{E},6~{R},10~{S})-6,10-bis(oxidanyl)undec-1-enyl]-4,6-bis(oxidanyl)benzoic acid, GLYCEROL, POTASSIUM ION, ... | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
5UAD
 
 | |
5JW6
 
 | |
1F73
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM III IN COMPLEX WITH SIALIC ACID ALDITOL | | Descriptor: | 2,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDO NONANOIC ACID, GLYCEROL, N-ACETYL NEURAMINATE LYASE | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-25 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
6JC6
 
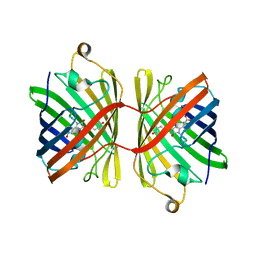 | |
6JH5
 
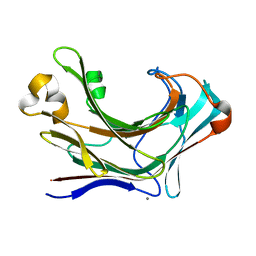 | | Structure of Marine bacterial laminarinase | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Ru, L, Tanokura, M, Long, L. | | Deposit date: | 2019-02-17 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JER
 
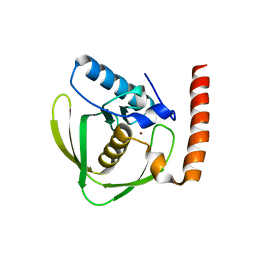 | | Apo crystal structure of class I type a peptide deformylase from Acinetobacter baumannii | | Descriptor: | Peptide deformylase, ZINC ION | | Authors: | Ho, T.H, Lee, I.H, Kang, L.W. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, crystallization, and preliminary X-ray crystallographic analysis of peptide deformylase from Acinetobacter baumanii
Biodesign, 5, 2017
|
|
