3F8M
 
 | | Crystal Structure of PhnF from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, GntR-family protein transcriptional regulator | | Authors: | Busby, J.N, Gebhard, S, Cook, G.M, Lott, S.J, Baker, E.N, Money, V.A. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PhnF, a GntR-family transcription regulator in Mycobacterium smegmatis
To be Published
|
|
3F74
 
 | | Crystal structure of wild type LFA1 I domain | | Descriptor: | GLYCEROL, Integrin alpha-L, MAGNESIUM ION | | Authors: | Zhang, H, Wang, J.-H. | | Deposit date: | 2008-11-07 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of isoflurane bound to integrin LFA-1 supports a unified mechanism of volatile anesthetic action in the immune and central nervous systems.
Faseb J., 23, 2009
|
|
3P62
 
 | |
3F92
 
 | | Crystal structure of ubiquitin-conjugating enzyme E2-25kDa (Huntington Interacting Protein 2) M172A mutant crystallized at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Wilson, R.C, Hughes, R.C, Flatt, J.W, Meehan, E.J, Ng, J.D, Twigg, P.D. | | Deposit date: | 2008-11-13 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3P6M
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans | | Descriptor: | ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans
To be Published
|
|
3F7S
 
 | |
3F8F
 
 | | Crystal structure of multidrug binding transcriptional regulator LmrR complexed with Daunomycin | | Descriptor: | DAUNOMYCIN, Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | Descriptor: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3FAT
 
 | | X-ray structure of iGluR4 flip ligand-binding core (S1S2) in complex with (S)-AMPA at 1.90A resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, ACETIC ACID, GLYCEROL, ... | | Authors: | Kasper, C, Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2008-11-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of agonist recognition by the ligand-binding core of the ionotropic glutamate receptor 4
Febs Lett., 582, 2008
|
|
3PA4
 
 | | X-ray crystal structure of compound 2a bound to human CHK1 kinase domain | | Descriptor: | 2-(4-chlorophenyl)-4-[(3S)-piperidin-3-ylamino]thieno[3,2-c]pyridine-7-carboxamide, GLYCEROL, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3FCL
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-{[(4-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}butyl)amino]methyl}benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3FCJ
 
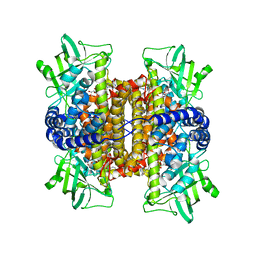 | | Nitroalkane oxidase: mutant402N crystallized with nitroethane | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Nitroalkane oxidase, ... | | Authors: | Major, D.T, Gao, J, Heroux, A, Orville, A.M, Valley, M.P, Fitzpatrick, P.F. | | Deposit date: | 2008-11-21 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Differential quantum tunneling contributions in nitroalkane oxidase catalyzed and the uncatalyzed proton transfer reaction.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3PEB
 
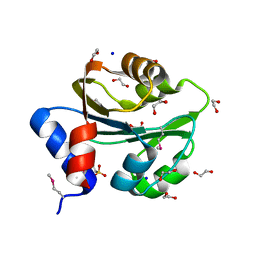 | | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Dipeptidase, ... | | Authors: | Cuff, M.E, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus.
TO BE PUBLISHED
|
|
3FDX
 
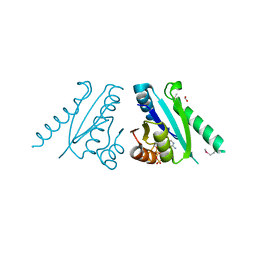 | | Putative filament protein / universal stress protein F from Klebsiella pneumoniae. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | X-ray crystal structure of putative filament protein / universal stress protein F from Klebsiella pneumoniae.
To be Published
|
|
3PEJ
 
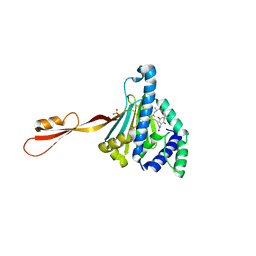 | | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of Macbecin | | Descriptor: | Endoplasmin homolog, MACBECIN, SULFATE ION | | Authors: | Wernimont, A.K, Tempel, W, Hutchinson, A, Weadge, J, MacKenzie, F, Senisterra, G, Vedadi, M, Cossar, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Pizzaro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of Macbecin
To be Published
|
|
3FCY
 
 | |
3PC4
 
 | |
3FE7
 
 | |
3PDJ
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with 4,4-Disubstituted Cyclohexylbenzamide Inhibitor | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-[trans-4-(3-amino-3-oxopropyl)-4-phenylcyclohexyl]-N-cyclopropyl-4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]benzamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Optimization of Novel 4,4-Disubstituted Cyclohexylbenzamide Derivatives as Potent 11beta-HSD1 Inhibitors
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3FDT
 
 | | Crystal structure of the complex of human chromobox homolog 5 (CBX5) with H3K9(me)3 peptide | | Descriptor: | Chromobox protein homolog 5, H3K9(me)3 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
3PE2
 
 | | Crystal structure of human protein kinase CK2 in complex with the inhibitor CX-5011 | | Descriptor: | 5-[(3-ethynylphenyl)amino]pyrimido[4,5-c]quinoline-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Battistutta, R, Papinutto, E, Lolli, G, Pierre, F, Haddach, M, Ryckman, D.M. | | Deposit date: | 2010-10-25 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer.
Biochemistry, 50, 2011
|
|
3FFS
 
 | | The Crystal Structure of Cryptosporidium parvum Inosine-5'-Monophosphate Dehydrogenase | | Descriptor: | Inosine-5-monophosphate dehydrogenase | | Authors: | Riera, T.V, D'Aquino, J.A, Lu, J, Petsko, G.A, Hedstrom, L. | | Deposit date: | 2008-12-04 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The structural basis of Cryptosporidium -specific IMP dehydrogenase inhibitor selectivity
J.Am.Chem.Soc., 132, 2010
|
|
3FGT
 
 | | Two chain form of the 66.3 kDa protein from mouse lacking the linker peptide | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
3IJU
 
 | |
3PDC
 
 | |
