4KLM
 
 | | DNA polymerase beta matched product complex with Mg2+, 11 h | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
6VB1
 
 | | HLA-B*15:02 complexed with a synthetic peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Schutte, R.J, Li, D, Andring, J, McKenna, R, Ostrov, D.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | HLA-B*15:02 complexed with a synthetic peptide
To Be Published
|
|
6VB7
 
 | |
6VBI
 
 | | crystal structure of PDE5 in complex with a non-competitive inhibitor | | Descriptor: | (13bS)-4,9-dimethoxy-14-methyl-8,13,13b,14-tetrahydroindolo[2',3':3,4]pyrido[2,1-b]quinazolin-5(7H)-one, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Ke, H, Luo, H.B. | | Deposit date: | 2019-12-18 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.30000758 Å) | | Cite: | Identification of a novel allosteric pocket and its regulation mechanism
To Be Published
|
|
8VR6
 
 | | crystal structure of the Pcryo_0619 N-acetryltransferase from Psychrobacter cryohalolentis K5 in the presence of CoA-disulfide | | Descriptor: | Acetyltransferase, putative, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Dunsirn, M.M, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
4KLG
 
 | | DNA polymerase beta matched product complex with Mg2+, 40 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
2M9Q
 
 | |
6YBL
 
 | | Structure of MBP-Mcl-1 in complex with compound 9m | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy]phenyl]propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
2MA5
 
 | | Solution NMR structure of PHD type Zinc finger domain of Lysine-specific demethylase 5B (PLU-1/JARID1B) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7375C | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Hassan, F, Ramelot, T.A, Yang, Y, Cort, J.R, Janjua, H, Kohan, E, Lee, D, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-28 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of PHD type Zinc finger domain of Lysine-specific
demethylase 5B (PLU-1/JARID1B) from Homo sapiens, Northeast Structural
Genomics Consortium (NESG) Target HR7375C
To be Published
|
|
2MB4
 
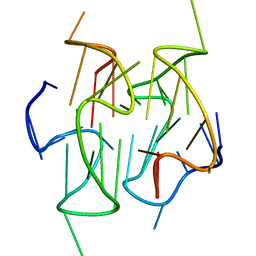 | | Solution structure of a stacked dimeric G-quadruplex formed by a segment of the human CEB1 minisatellite | | Descriptor: | DNA_(5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*TP*GP*G)-3') | | Authors: | Adrian, M, Ang, D.J, Lech, C, Heddi, B, Nicolas, A, Phan, A.T. | | Deposit date: | 2013-07-25 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Conformational Dynamics of a Stacked Dimeric G-Quadruplex Formed by the Human CEB1 Minisatellite.
J.Am.Chem.Soc., 136, 2014
|
|
4TKL
 
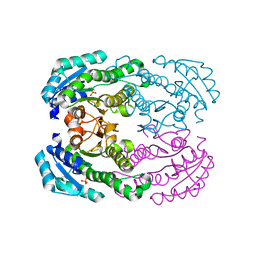 | | Crystal structure of NADH-dependent reductase A1-R' responsible for alginate metabolism | | Descriptor: | NADH-dependent reductase for 4-deoxy-L-erythro-5-hexoseulose uronate, PHOSPHATE ION | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
2MES
 
 | |
4KQ5
 
 | |
8OLW
 
 | |
4RT6
 
 | | Structure of a complex between hemopexin and hemopexin binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heme/hemopexin-binding protein, Hemopexin | | Authors: | Zambolin, S, Clantin, B, Haouz, A, Villeret, V, Delepelaire, P. | | Deposit date: | 2014-11-13 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for haem piracy from host haemopexin by Haemophilus influenzae.
Nat Commun, 7, 2016
|
|
4K7V
 
 | | OYE1-W116A complexed with (R)-carvone | | Descriptor: | (5R)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-one, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sullivan, B, Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2013-04-17 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the
Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
5HEK
 
 | | crystal structure of M1.HpyAVI | | Descriptor: | Adenine specific DNA methyltransferase (DpnA) | | Authors: | Ma, B, Zhang, H, Liu, W. | | Deposit date: | 2016-01-06 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical and structural characterization of a DNA N6-adenine methyltransferase from Helicobacter pylori
Oncotarget, 7, 2016
|
|
8OLS
 
 | |
4KA7
 
 | | Structure of Organellar OligoPeptidase (E572Q) in complex with an endogenous substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Oligopeptidase A, ... | | Authors: | Berntsson, R.P.-A, Kmiec, B, Teixeira, P.F, Svensson, L.M, Bakali, A, Glaser, E, Stenmark, P. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
3CWO
 
 | | A beta/alpha-barrel built by the combination of fragments from different folds | | Descriptor: | SULFATE ION, beta/alpha-barrel protein based on 1THF and 1TMY | | Authors: | Bharat, T.A.M, Eisenbeis, S, Zeth, K, Hocker, B. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A beta alpha-barrel built by the combination of fragments from different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8OM0
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Na+, Mg2+ and intronistat B | | Descriptor: | 3,4,5-trihydroxybenzoic acid, Domains 1-5, MAGNESIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2023-03-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
3EPR
 
 | | Crystal structure of putative HAD superfamily hydrolase from Streptococcus agalactiae. | | Descriptor: | GLYCEROL, Hydrolase, haloacid dehalogenase-like family, ... | | Authors: | Ramagopal, U.A, Toro, R, Dickey, M, Tang, B.K, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of putative HAD superfamily hydrolase from Streptococcus agalactiae.
To be Published
|
|
5HI1
 
 | | Backbone Modifications in the Protein GB1 Helix: Aib24, beta-3-Lys28, beta-3-Lys31, Aib35 | | Descriptor: | ACETATE ION, Immunoglobulin G-binding protein G | | Authors: | Tavenor, N.A, Reinert, Z.E, Lengyel, G.A, Griffith, B.D, Horne, W.S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold.
Chem.Commun.(Camb.), 52, 2016
|
|
2MGN
 
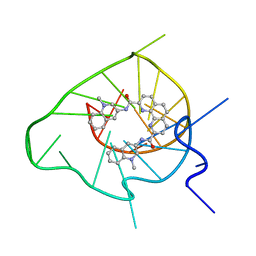 | | Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC3 | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3', N2,N9-bis(1-methylquinolin-3-yl)-1,10-phenanthroline-2,9-dicarboxamide | | Authors: | Chung, W.J, Heddi, B, Hamon, F, Teulade-Fichou, M.P, Phan, A.T. | | Deposit date: | 2013-11-01 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a G-quadruplex Bound to the Bisquinolinium Compound Phen-DC3.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3ERV
 
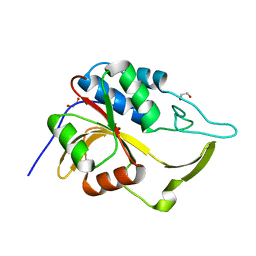 | | Crystal structure of an putative C39-like peptidase from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, putative C39-like peptidase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an putative C39-like peptidase from Bacillus anthracis
To be Published
|
|
