2O7A
 
 | | T4 lysozyme C-terminal fragment | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme | | Authors: | Echols, N, Kwon, E, Marqusee, S.M, Alber, T. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
4EIC
 
 | |
7BNH
 
 | |
7WKG
 
 | | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with erucic acid | | Descriptor: | (Z)-docos-13-enoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with erucic acid
To Be Published
|
|
7G0Z
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with 5-[[3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]-3,6-dihydro-2H-pyran-4-carboxylic acid | | Descriptor: | 5-{[(3M)-3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}-3,6-dihydro-2H-pyran-4-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Crystal Structure of a human FABP4 binding site mutated to that of FABP5 complex
To be published
|
|
7TX0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 9 (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-02-07 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 9 (P43 crystal form)
To Be Published
|
|
7FFK
 
 | | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitoleic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-23 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitoleic acid
To Be Published
|
|
1XVO
 
 | | Trypsin from Fusarium oxysporum at pH 6 | | Descriptor: | SULFATE ION, trypsin | | Authors: | Schmidt, A, Lamzin, V.S. | | Deposit date: | 2004-10-28 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Extraction of functional motion in trypsin crystal structures.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3ZQV
 
 | |
1P9G
 
 | | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Ecommia ulmoides Oliver at atomic resolution | | Descriptor: | ACETATE ION, EAFP 2 | | Authors: | Xiang, Y, Huang, R.H, Liu, X.Z, Wang, D.C. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Eucommia ulmoides Oliver at an atomic resolution.
J.Struct.Biol., 148, 2004
|
|
2HS1
 
 | | Ultra-high resolution X-ray crystal structure of HIV-1 protease V32I mutant with TMC114 (darunavir) inhibitor | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weber, I.T, Kovalevsky, A.Y. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Ultra-high Resolution Crystal Structure of HIV-1 Protease Mutant Reveals Two Binding Sites for Clinical Inhibitor TMC114.
J.Mol.Biol., 363, 2006
|
|
4LAU
 
 | | Crystal structure of human AR complexed with NADP+ and {2-[(4-bromobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-bromobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-20 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.843 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
4PSS
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 100K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.849 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4O8H
 
 | | 0.85A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
7P4R
 
 | | Ultra High Resolution X-ray Structure of Orthorhombic Bovine Pancreatic Ribonuclease at 100K | | Descriptor: | ETHANOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Lisgarten, D.R, Palmer, R.A, Cooper, J.B, Naylor, C.E, Howlin, B.J, Lisgarten, J.N, Najmudin, S, Lobley, C.M.C. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ultra-high resolution X-ray structure of orthorhombic bovine pancreatic Ribonuclease A at 100K.
BMC Chem, 17, 2023
|
|
4AYO
 
 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
5EMB
 
 | |
7RVE
 
 | |
6AIR
 
 | | High resolution structure of perdeuterated high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
4D5M
 
 | | Gonadotropin-releasing hormone agonist | | Descriptor: | PHOSPHATE ION, TRIPTORELIN | | Authors: | Legrand, P, Le Du, M.-H, Valery, C, Deville-Foillard, S, Paternostre, M, Artzner, F. | | Deposit date: | 2014-11-05 | | Release date: | 2015-08-12 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Atomic View of the Histidine Environment Stabilizing Higher- Ph Conformations of Ph-Dependent Proteins.
Nat.Commun., 6, 2015
|
|
6AIQ
 
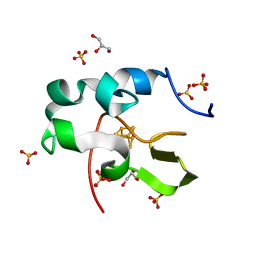 | | High resolution structure of recombinant high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
4XXG
 
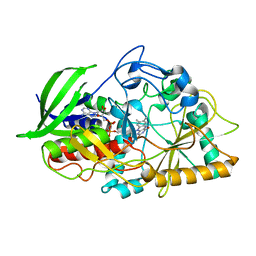 | |
1X8Q
 
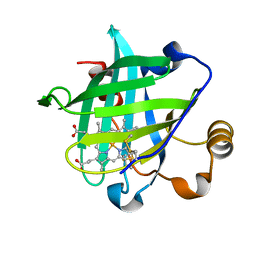 | | 0.85 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus in Complex with Water at pH 5.6 | | Descriptor: | Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
5XVT
 
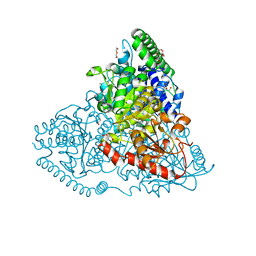 | | Crystal Structure of Transketolase in complex with hydroxylated TPP from Pichia Stipitis, crystal 2 | | Descriptor: | 2-[(2R)-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-oxidanyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Mesomeric Effect of Thiazolium on non-Kekule Diradicals in Pichia stipitis Transketolase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6ETL
 
 | |
