5FXY
 
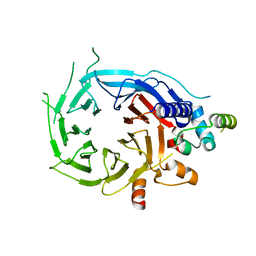 | | Structure of the human RBBP4:MTA1(464-546) complex | | Descriptor: | HISTONE-BINDING PROTEIN RBBP4, METASTASIS-ASSOCIATED PROTEIN MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
6MQF
 
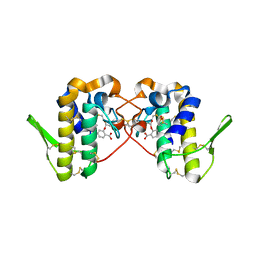 | |
6G86
 
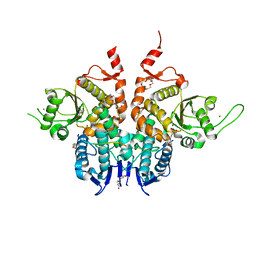 | | Structure of Cdc14 bound to SIC1 PxL motif | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Mouilleron, S, Kataria, M, Uhlmann, F. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A PxL motif promotes timely cell cycle substrate dephosphorylation by the Cdc14 phosphatase.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6Y7O
 
 | | The complex between the eight-bladed symmetrical designer protein Tako8 and the silicotungstic acid Keggin (STA) | | Descriptor: | Keggin (STA), Tako8 | | Authors: | Vandebroek, L, Noguchi, H, Parac-Vogt, T.N, Van Meervelt, L, Voet, A.R.D. | | Deposit date: | 2020-03-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Shape and Size Complementarity-Induced Formation of Supramolecular Protein Assemblies with Metal-Oxo Clusters
Cryst.Growth Des., 2021
|
|
5DZL
 
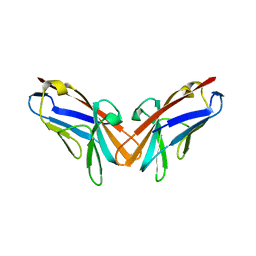 | | Crystal structure of the protein human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Huang, Y.H, Russell, A, Gandhi, A.K, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
6POT
 
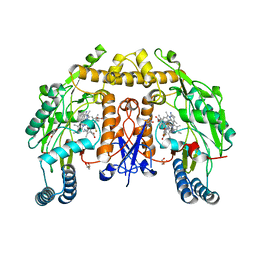 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-(thiazol-5-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(1,3-thiazol-5-yl)methoxy]phenyl}-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5DZR
 
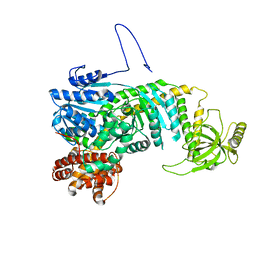 | |
5L4H
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with 5-(2-BROMO-ETHYL)-5-ETHYL-PYRIMIDINE-2,4,6-TRIONE (brominated barbiturate) | | Descriptor: | 5-(2-bromoethyl)-5-ethyl-1,3-diazinane-2,4,6-trione, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Ruza, R.R, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
5VDT
 
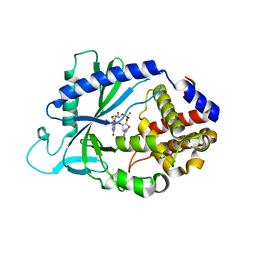 | | Human cyclic GMP-AMP synthase (cGAS) in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Byrnes, L.J, Hall, J.D. | | Deposit date: | 2017-04-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition.
Protein Sci., 26, 2017
|
|
5G03
 
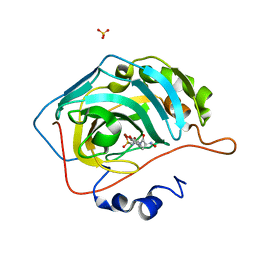 | | An unusual natural product primary sulfonamide: synthesis, carbonic anhydrase inhibition and protein x-ray structure of Psammaplin C | | Descriptor: | CARBONIC ANHYDRASE 2, PSAMMAPLIN C, SULFATE ION, ... | | Authors: | Mujumdar, P, Supuran, C.T, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Unusual Natural Product Primary Sulfonamide: Synthesis, Carbonic Anhydrase Inhibition and Protein X-Ray Structures of Psammaplin C.
J.Med.Chem., 59, 2016
|
|
6PIC
 
 | |
5L59
 
 | | Plexin A1 full extracellular region, domains 1 to 10, to 6 angstrom, spacegroup P2(1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5G0S
 
 | | InhA in complex with a DNA encoded library hit | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], N-[4-[2-[(2S)-4-[2-(methylamino)-2-oxidanylidene-ethyl]-3-oxidanylidene-2-(phenylmethyl)piperazin-1-yl]-2-oxidanylidene-ethyl]cyclohexyl]-2-(3-methyl-1-benzothiophen-2-yl)ethanamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Cofactor-Specific, Bactericidal Mycobacterium Tuberculosis Inha Inhibitors Using DNA-Encoded Library Technology
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5G17
 
 | | Bordetella Alcaligenes HDAH (T101A) bound to 9,9,9-trifluoro-8,8- dihydroxy-N-phenylnonanamide. | | Descriptor: | 9,9,9-tris(fluoranyl)-8,8-bis(oxidanyl)-~{N}-phenyl-nonanamide, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
6PP0
 
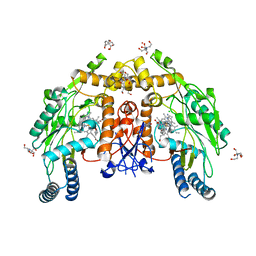 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-(cyclobutylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-(cyclobutylmethoxy)phenyl]-4-methylquinolin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5E74
 
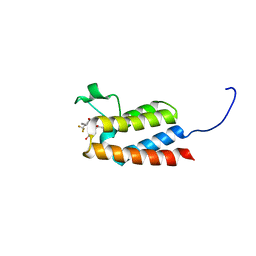 | | Crystal Structure of BAZ2B bromodomain in complex with acetylindole compound UZH50 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)-3-(trifluoromethyl)benzamide | | Authors: | Lolli, G, Spiliotopoulos, D, Dolbois, A, Nevado, C, Caflisch, A. | | Deposit date: | 2015-10-11 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The "Gatekeeper" Residue Influences the Mode of Binding of Acetyl Indoles to Bromodomains.
J. Med. Chem., 59, 2016
|
|
5VF1
 
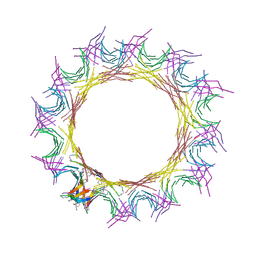 | |
5E0H
 
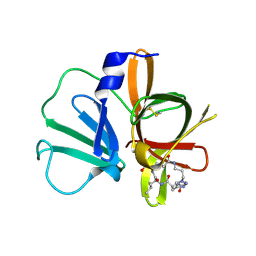 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic (18-mer) inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(9~{S},12~{S},15~{S})-9-(hydroxymethyl)-12-(2-methylpropyl)-6,11,14-tris(oxidanylidene)-1,5,10,13,18,19-hexazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate, GLYCEROL, Norovirus 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Weerawarna, P.M, Kim, Y, Kankanamalage, A.C.G, Damalanka, V.C, Lushington, G.H, Alliston, K.R, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-09-28 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies.
Eur.J.Med.Chem., 119, 2016
|
|
8OK9
 
 | |
5KU5
 
 | |
5FK5
 
 | |
6MUJ
 
 | | Formylglycine generating enzyme bound to copper | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Lafrance-Vanasse, J, Appel, M.J, Tsai, C.-L, Bertozzi, C, Tainer, J.A. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Formylglycine-generating enzyme binds substrate directly at a mononuclear Cu(I) center to initiate O2activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6G9D
 
 | |
6Y5T
 
 | | Crystal structure of savinase at room temperature | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
6FUB
 
 | |
