6MLT
 
 | | Crystal structure of the V. cholerae biofilm matrix protein Bap1 | | Descriptor: | CALCIUM ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Kaus, K, Biester, A, Chupp, E, Lu, K, Vidsudharomn, C, Olson, R. | | Deposit date: | 2018-09-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 angstrom crystal structure of the extracellular matrix protein Bap1 fromVibrio choleraeprovides insights into bacterial biofilm adhesion.
J.Biol.Chem., 294, 2019
|
|
7CFJ
 
 | |
6GDQ
 
 | |
5VI4
 
 | | IL-33/ST2/IL-1RAcP ternary complex structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein, Interleukin-1 receptor-like 1, ... | | Authors: | Guenther, S, Sundberg, E.J. | | Deposit date: | 2017-04-13 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | IL-1 Family Cytokines Use Distinct Molecular Mechanisms to Signal through Their Shared Co-receptor.
Immunity, 47, 2017
|
|
6URW
 
 | | Crystal structure of ricin A chain in complex with inhibitor 4-(2-thienylmethyl)benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[(thiophen-2-yl)methyl]benzoic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Harijan, R.K, Li, X.P, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small Molecule Inhibitors Targeting the Interaction of Ricin Toxin A Subunit with Ribosomes.
Acs Infect Dis., 6, 2020
|
|
7CG9
 
 | |
6PNG
 
 | |
6GDV
 
 | | Structure of CutA from Synechococcus elongatus PCC7942 complexed with Bis-Tris molecule | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
6US0
 
 | | Barrier-to-autointegration factor soaked in R,S,R-bisfuranol (RSR): 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US8
 
 | |
8OQO
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-49 | | Descriptor: | GLYCEROL, Probable fatty oxidation protein FadB, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6PO9
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-(pyridin-2-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(pyridin-2-yl)methoxy]phenyl}-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-03 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5VJP
 
 | | Crystal Structure of Adenine Phosphoribosyltransferase from Saccharomyces cerevisiae Complexed with L-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (L-DIAB) and Adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Adenine phosphoribosyltransferase 1, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Synthesis of bis-Phosphate Iminoaltritol Enantiomers and Structural Characterization with Adenine Phosphoribosyltransferase.
ACS Chem. Biol., 13, 2018
|
|
6G27
 
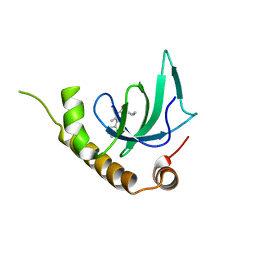 | | X-ray structure of NSD3-PWWP1 in complex with compound 5 | | Descriptor: | 5-methyl-6-phenyl-2-piperidin-4-yl-pyridazin-3-one, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
8OMK
 
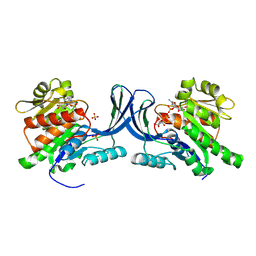 | | hKHK-C in complex with ADP & fructose 1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, ... | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
6UTE
 
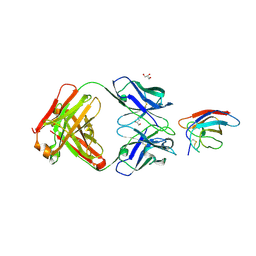 | | Crystal structure of Z032 Fab in complex with WNV EDIII | | Descriptor: | Envelope domain III, GLYCEROL, Z032 Fab heavy chain, ... | | Authors: | Esswein, S.R, Gristick, H.B, Keeffe, J.R, Bjorkman, P.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YQ6
 
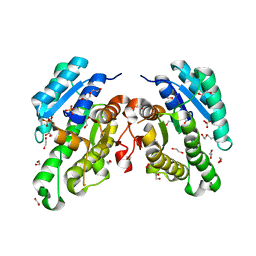 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6VH0
 
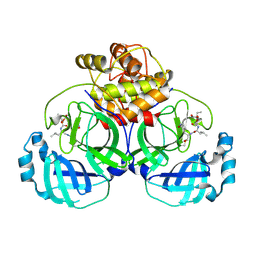 | | 1.95 A resolution structure of MERS 3CL protease in complex with inhibitor 6g | | Descriptor: | N~2~-{[(5-ethyl-1,3-dioxan-5-yl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
8OE6
 
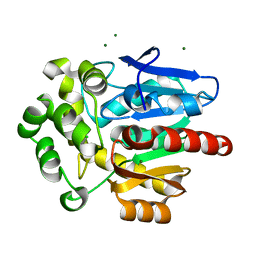 | | Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
6G4Y
 
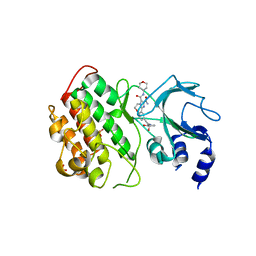 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 1a | | Descriptor: | 10-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrolidin-3-yl]ethynyl]-~{N}3-(oxan-4-yl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Hole, A.J, Hymowitz, S.G, McEwan, P.A. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
5KR1
 
 | | Protease PR5-DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease PR5-DRV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5HA7
 
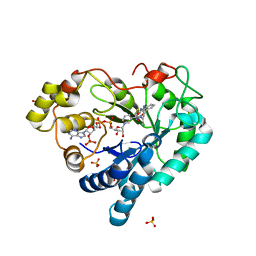 | | Human Aldose Reductase in Complex with NADP+ and WY14643 in Space Group P212121 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sawaya, M.R, Cascio, D, Balendiran, G.K. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of WY 14,643 and its Complex with Aldose Reductase.
Sci Rep, 6, 2016
|
|
6FOA
 
 | | Human Xylosyltransferase 1 apo structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E. | | Deposit date: | 2018-02-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | Structural Basis for the Initiation of Glycosaminoglycan Biosynthesis by Human Xylosyltransferase 1.
Structure, 26, 2018
|
|
8OFK
 
 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under reducing conditions (space group C 2 2 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, CHLORIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.713 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
6FOP
 
 | | Glycoside hydrolase family 81 from Clostridium thermocellum (CtLam81A), Mutant E515A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Correia, M.A.S.C, Carvalho, A.L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel insights into the degradation of beta-1,3-glucans by the cellulosome of Clostridium thermocellum revealed by structure and function studies of a family 81 glycoside hydrolase.
Int.J.Biol.Macromol., 117, 2018
|
|
