8HI4
 
 | |
1U9Q
 
 | | Crystal structure of cruzain bound to an alpha-ketoester | | Descriptor: | [1-(1-METHYL-4,5-DIOXO-PENT-2-ENYLCARBAMOYL)-2-PHENYL-ETHYL]-CARBAMIC ACID BENZYL ESTER, cruzipain | | Authors: | Lange, M, Weston, S.G, Cheng, H, Culliane, M, Fiorey, M.M, Grisostomi, C, Hardy, L.W, Hartstough, D.S, Pallai, P.V, Tilton, R.F, Baldino, C.M, Brinen, L.S, Engel, J.C, Choe, Y, Price, M.S, Craik, C.S. | | Deposit date: | 2004-08-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of alpha-keto-based inhibitors of cruzain, a cysteine protease implicated in Chagas disease
Bioorg.Med.Chem., 13, 2005
|
|
6Z0W
 
 | |
8HI8
 
 | |
8HCI
 
 | |
8H2D
 
 | |
5G5T
 
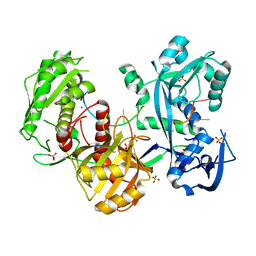 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii in complex with guide DNA | | Descriptor: | ARGONAUTE, GUIDE DNA, MAGNESIUM ION, ... | | Authors: | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | Deposit date: | 2016-06-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
8HI7
 
 | |
2ZTM
 
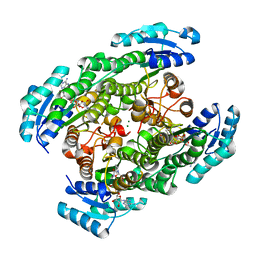 | | T190S mutant of D-3-hydroxybutyrate dehydrogenase | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-07 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
6ZIQ
 
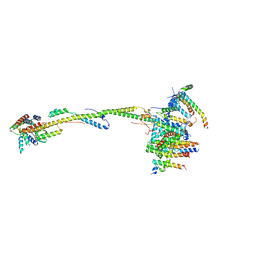 | | bovine ATP synthase stator domain, state 1 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5DRJ
 
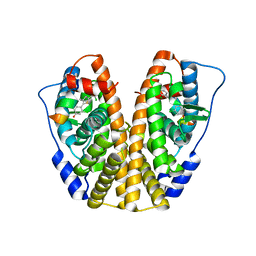 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a dichloro-substituted, 3-methyl 2,5-diarylthiophene-core ligand 4,4'-(3-methylthiene-2,5-diyl)bis(3-chlorophenol) | | Descriptor: | 4,4'-(3-methylthiene-2,5-diyl)bis(3-chlorophenol), Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
1DUP
 
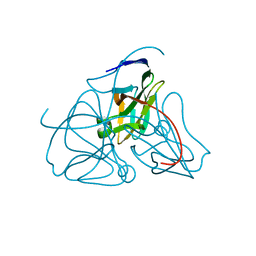 | | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE (D-UTPASE) | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Wilson, K.S, Larsson, G, Nyman, P.O, Cedergren, E. | | Deposit date: | 1995-09-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a dUTPase.
Nature, 355, 1992
|
|
5G5S
 
 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGONAUTE, MAGNESIUM ION | | Authors: | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | Deposit date: | 2016-06-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
8H0H
 
 | |
8GXN
 
 | | The crystal structure of CsFAOMT2 in complex with SAH | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, caffeyl-CoA-O-methyltransferase | | Authors: | Zhang, Z.M, Zhou, Y.E, Huang, H.S. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Characterization of two O-methyltransferases involved in the biosynthesis of O-methylated catechins in tea plant.
Nat Commun, 14, 2023
|
|
2DSU
 
 | | Binding of chitin-like polysaccharide to protective signalling factor: Crystal structure of the complex formed between signalling protein from sheep (SPS-40) with a tetrasaccharide at 2.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Kaur, P, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
2DSQ
 
 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 1, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6ZG7
 
 | | bovine ATP synthase rotor domain, state 1 | | Descriptor: | ATP synthase F(0) complex subunit C2, mitochondrial, ATP synthase subunit delta, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZIT
 
 | | bovine ATP synthase Stator domain, state 2 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C2, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2IKI
 
 | | Human aldose reductase complexed with halogenated IDD-type inhibitor | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2006-10-02 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Thermodynamic Study on Aldose Reductase: Nitro-substituted Inhibitors with Strong Enthalpic Binding Contribution
J.Mol.Biol., 368, 2007
|
|
8GXO
 
 | |
6Z38
 
 | |
6Z37
 
 | |
1UH0
 
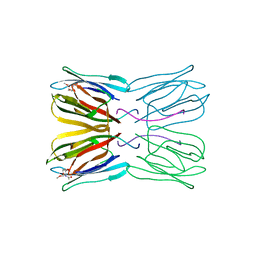 | | Crystal structure of jacalin- Me-alpha-GalNAc complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, methyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
4B1T
 
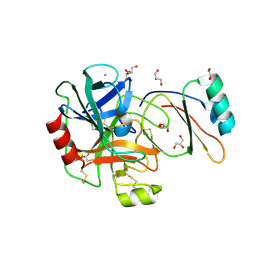 | | Structure of the factor Xa-like trypsin variant triple-Ala (TA) in complex with eglin C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Menzel, A, Neumann, P, Stubbs, M.T. | | Deposit date: | 2012-07-12 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Thermodynamic signatures in macromolecular interactions involving conformational flexibility.
Biol.Chem., 395, 2014
|
|
