7BPY
 
 | | X-ray structure of human PPARalpha ligand binding domain-clofibric acid-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-(4-chloranylphenoxy)-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BQ2
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, 15-meric peptide from Nuclear receptor coactivator 1, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5BRY
 
 | | HIV-1 wild Type protease with GRL-011-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, sulfonamide isostere derivate) | | Descriptor: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-06-01 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Design of HIV-1 Protease Inhibitors with Amino-bis-tetrahydrofuran Derivatives as P2-Ligands to Enhance Backbone-Binding Interactions: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Studies.
J.Med.Chem., 58, 2015
|
|
6QTN
 
 | | Tubulin-cyclostreptin complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Balaguer, F.d.A, Muehlethaler, T, Estevez-Gallego, J, Calvo, E, Gimenez-Abian, J.F, Risinger, A.L, Sorensen, E.J, Vanderwal, C.D, Altmann, K.-H, Mooberry, S.L, Steinmetz, M.O, Oliva, M.A, Prota, A.E, Diaz, J.F. | | Deposit date: | 2019-02-25 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Cyclostreptin-Tubulin Adduct: Implications for Tubulin Activation by Taxane-Site Ligands.
Int J Mol Sci, 20, 2019
|
|
7QAA
 
 | | Crystal structure of RARalpha/RXRalpha ligand binding domain heterodimer in complex with BMS614 and oleic acid | | Descriptor: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, Isoform Alpha-1-deltaBC of Retinoic acid receptor alpha, OLEIC ACID, ... | | Authors: | le Maire, A, Vivat, V, Guee, L, Blanc, P, Malosse, C, Chamot-Rooke, J, Germain, P, Bourguet, w. | | Deposit date: | 2021-11-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
2P4Y
 
 | | Crystal structure of human PPAR-gamma-ligand binding domain complexed with an indole-based modulator | | Descriptor: | (2R)-2-(4-CHLORO-3-{[3-(6-METHOXY-1,2-BENZISOXAZOL-3-YL)-2-METHYL-6-(TRIFLUOROMETHOXY)-1H-INDOL-1-YL]METHYL}PHENOXY)PROPANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | McKeever, B.M. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The differential interactions of peroxisome proliferator-activated receptor gamma ligands with Tyr473 is a physical basis for their unique biological activities.
Mol.Pharmacol., 73, 2008
|
|
6GFX
 
 | | pVHL:EloB:EloC in complex with modified HIF-1a CODD peptide containing (3R,4S)-3-fluoro-4-hydroxyproline (ligand 13a) | | Descriptor: | Elongin-B, Elongin-C, FLUORINATED HYPOXIA-INDUCIBLE FACTOR 1 ALPHA PEPTIDE, ... | | Authors: | Castro, G.V, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
7PDQ
 
 | | Crystal structure of a mutated form of RXRalpha ligand binding domain in complex with LG100268 and a coactivator fragment | | Descriptor: | 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W, Guee, L. | | Deposit date: | 2021-08-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
7WPM
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPN
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor and ATP | | Descriptor: | (phenylmethyl) N-[(2R)-2-[2-(4-bromanyl-3-oxidanyl-phenyl)-5-(methylcarbamoyl)benzimidazol-1-yl]-2-(3,4-dimethoxyphenyl)ethyl]carbamate, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
1MZN
 
 | | CRYSTAL STRUCTURE at 1.9 ANGSTROEMS RESOLUTION OF THE HOMODIMER OF HUMAN RXR ALPHA LIGAND BINDING DOMAIN BOUND TO THE SYNTHETIC AGONIST COMPOUND BMS 649 AND A COACTIVATOR PEPTIDE | | Descriptor: | 4-[2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-[1,3]DIOXOLAN-2-YL]-BENZOIC ACID, Nuclear receptor coactivator 2, RXR retinoid X receptor | | Authors: | Egea, P.F, Mitschler, A, Moras, D. | | Deposit date: | 2002-10-09 | | Release date: | 2002-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Recognition of Agonist Ligands by RXRs
MOL.ENDOCRINOL., 16, 2002
|
|
1OW0
 
 | | Crystal structure of human FcaRI bound to IgA1-Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ig alpha-1 chain C region, Immunoglobulin alpha Fc receptor, ... | | Authors: | Herr, A.B, Ballister, E.R, Bjorkman, P.J. | | Deposit date: | 2003-03-27 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Insights into IgA-mediated immune responses from the crystal structures of human Fc-alpha-RI and its complex with IgA1-Fc
Nature, 423, 2003
|
|
6QMU
 
 | |
7OKQ
 
 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | Descriptor: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | Authors: | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
4N73
 
 | |
4KB9
 
 | | Crystal structure of wild-type HIV-1 protease with novel tricyclic P2-ligands GRL-0739A | | Descriptor: | (3aR,3bR,4S,7aR,8aS)-decahydrofuro[2,3-b][1]benzofuran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2013-04-23 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Highly Potent HIV-1 Protease Inhibitors with Novel Tricyclic P2 Ligands: Design, Synthesis, and Protein-Ligand X-ray Studies.
J.Med.Chem., 56, 2013
|
|
4O3A
 
 | | Crystal structure of the glua2 ligand-binding domain in complex with L-aspartate at 1.80 a resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, F, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O3B
 
 | | Crystal structure of an open/closed glua2 ligand-binding domain dimer at 1.91 A resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krintel, C, de Rabassa, A.C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O3C
 
 | | Crystal structure of the GLUA2 ligand-binding domain in complex with L-aspartate at 1.50 A resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, K, Kaern, A.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
7WPJ
 
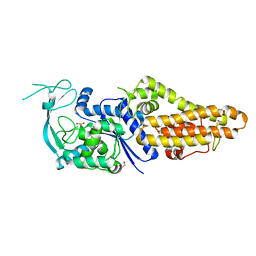 | | Methionyl-tRNA synthetase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
1AEF
 
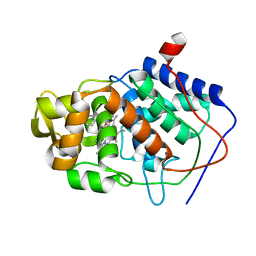 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (3-AMINOPYRIDINE) | | Descriptor: | 3-AMINOPYRIDINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1QWF
 
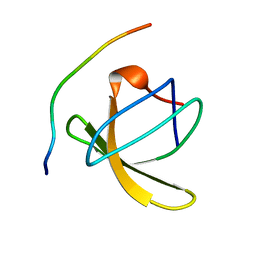 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND VSL12 | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC, VAL-SER-LEU-ALA-ARG-ARG-PRO-LEU-PRO-PRO-LEU-PRO | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1QWE
 
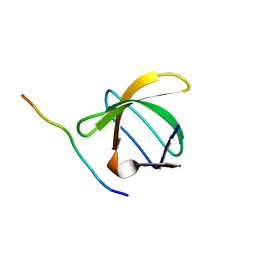 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND APP12 | | Descriptor: | ALA-PRO-PRO-LEU-PRO-PRO-ARG-ASN-ARG-PRO-ARG-LEU, TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1KVH
 
 | | NCSi-gb-bulge-DNA complex induced formation of a DNA bulge structure by a molecular wedge ligand-post-activated neocarzinostatin chromophore | | Descriptor: | 5'-D(*CP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*TP*TP*CP*GP*GP*G)-3', SPIRO[[7-METHOXY-5-METHYL-1,2-DIHYDRO-NAPHTHALENE]-3,1'-[5-HYDROXY-9-[2-METHYLAMINO-2,6-DIDEOXYGALACTOPYRANOSYL-OXY]-5-(2-OXO-[1,3]DIOXOLAN-4-YL)-3A,5,9,9A-TETRAHYDRO-3H-1-OXA-CYCLOPENTA[A]-S-INDACEN-2-ONE]] | | Authors: | Gao, X, Stassinopoulos, A, Ji, J, Kwon, Y, Bare, S, Goldberg, I.H. | | Deposit date: | 2002-01-26 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Induced formation of a DNA bulge structure by a molecular wedge ligand-postactivated neocarzinostatin chromophore.
Biochemistry, 41, 2002
|
|
1AEE
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (ANILINE) | | Descriptor: | ANILINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
