7Z1O
 
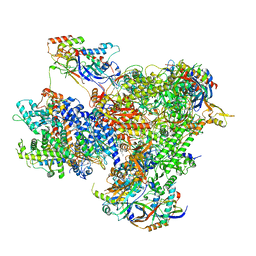 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1M
 
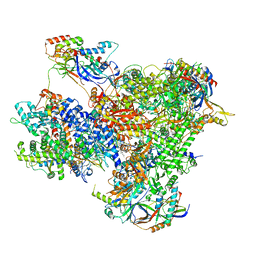 | | Structure of yeast RNA Polymerase III Elongation Complex (EC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
2G1Z
 
 | |
3MQ0
 
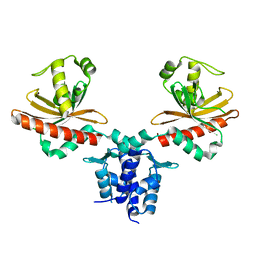 | | Crystal Structure of Agobacterium tumefaciens repressor BlcR | | Descriptor: | Transcriptional repressor of the blcABC operon | | Authors: | Chen, L. | | Deposit date: | 2010-04-27 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | The Agrobacterium tumefaciens Transcription Factor BlcR Is Regulated via Oligomerization.
J.Biol.Chem., 286, 2011
|
|
8WQ8
 
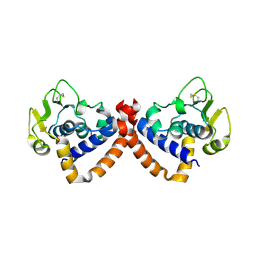 | |
8WQN
 
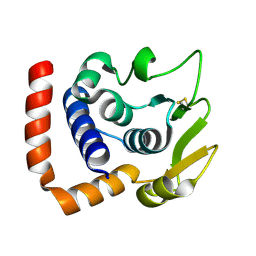 | |
8YK9
 
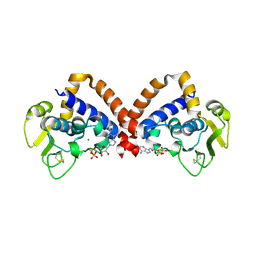 | | Structure of Saccharolobus solfataricus SegC (SSO0033) protein, ATP soak | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SegC | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2024-03-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unraveling the structure and function of a novel SegC protein interacting with the SegAB chromosome segregation complex in Archaea.
Nucleic Acids Res., 2024
|
|
5MRU
 
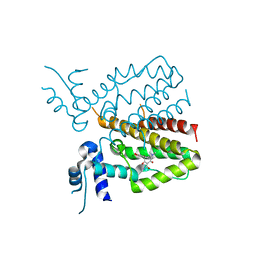 | |
4MQ9
 
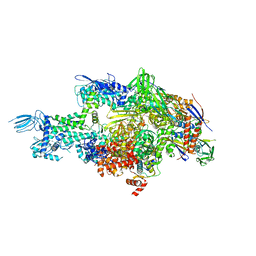 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme in complex with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ho, M.X, Arnold, E, Ebright, R.H, Zhang, Y, Tuske, S. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
1G6N
 
 | | 2.1 ANGSTROM STRUCTURE OF CAP-CAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | Passner, J.M, Schultz, S.C, Steitz, T.A. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution.
J.Mol.Biol., 304, 2000
|
|
5O1D
 
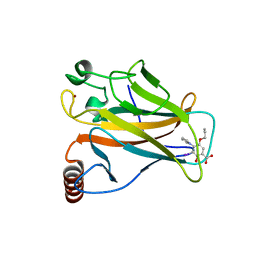 | | p53 cancer mutant Y220C in complex with compound MB481 | | Descriptor: | 3-iodanyl-2-oxidanyl-5-propoxy-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5XBT
 
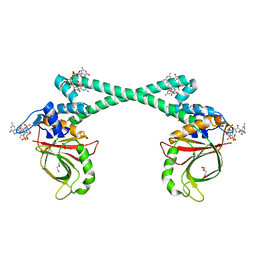 | | The structure of BrlR bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
6AWB
 
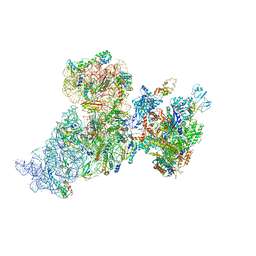 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWD
 
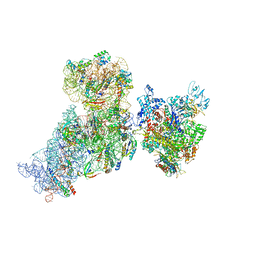 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWC
 
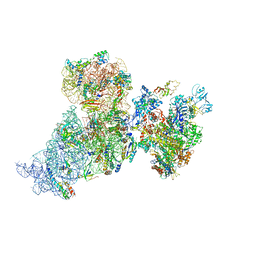 | | Structure of 30S ribosomal subunit and RNA polymerase complex in rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6X43
 
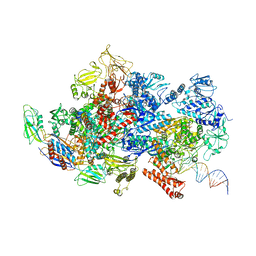 | | Mfd-bound E.coli RNA polymerase elongation complex - II state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
5XBW
 
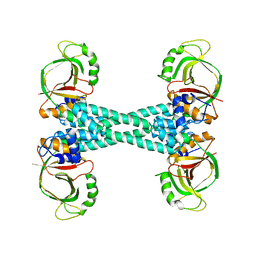 | | The structure of BrlR | | Descriptor: | Probable transcriptional regulator | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
4O61
 
 | | Structure of human ALKBH5 crystallized in the presence of citrate | | Descriptor: | CITRIC ACID, GLYCEROL, RNA demethylase ALKBH5, ... | | Authors: | Tempel, W, Chao, X, Liu, K, Dong, A, Cerovina, T, He, H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation.
J.Biol.Chem., 289, 2014
|
|
7B0Y
 
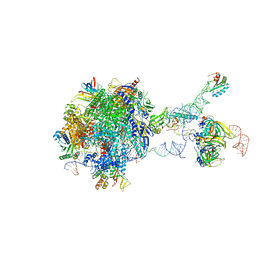 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
5FLV
 
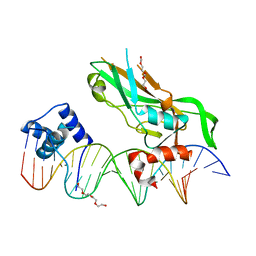 | | Crystal structure of NKX2-5 and TBX5 bound to the Nppa promoter region | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*TP*TP*CP*AP*AP*AP*GP*GP*TP *GP*TP*GP*AP*GP*AP*AP*G)-3', 5'-D(*TP*CP*TP*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP *TP*GP*AP*AP*GP*TP*GP*G)-3', HOMEOBOX PROTEIN NKX-2.5, ... | | Authors: | Stirnimann, C.U, Glatt, S, Mueller, C.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis.
Cell(Cambridge,Mass.), 164, 2016
|
|
5LM7
 
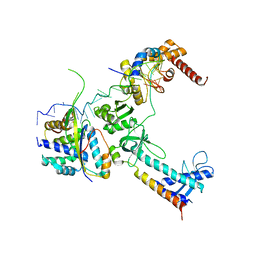 | | Crystal structure of the lambda N-Nus factor complex | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | Authors: | Said, N, Santos, K, Weber, G, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
7STE
 
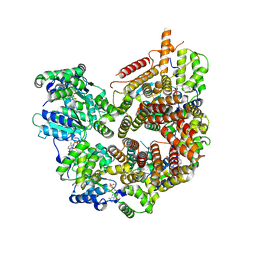 | | Rad24-RFC ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Checkpoint protein RAD24, ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6QVO
 
 | | Crystal structure of human MTH1 in complex with N6-methyl-dAMP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, N6-METHYL-DEOXY-ADENOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Scaletti, E, Vallin, K.S, Brautigam, L, Sarno, A, Warpman Berglund, U, Helleday, T, Stenmark, P, Jemth, A.S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | MutT homologue 1 (MTH1) removes N6-methyl-dATP from the dNTP pool.
J.Biol.Chem., 295, 2020
|
|
6XMI
 
 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Fab Heavy chain, Fab Light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7TAD
 
 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
