5M4V
 
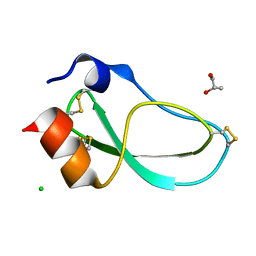 | | X-ray structure of the mambaquaretin-1, a selective antagonist of the vasopressin type 2 receptor | | Descriptor: | CHLORIDE ION, Mambaquaretin-1, S-1,2-PROPANEDIOL | | Authors: | Stura, E.A, Vera, L, Ciolek, J, Mourier, G, Gilles, N. | | Deposit date: | 2016-10-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7M17
 
 | | SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-1) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 7-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}heptanoic acid, Volume-regulated anion channel subunit LRRC8A | | Authors: | Kern, D.M, Gerber, E.E, Brohawn, S.G. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Small molecule SWELL1 complex induction improves glycemic control and nonalcoholic fatty liver disease in murine Type 2 diabetes.
Nat Commun, 13, 2022
|
|
7M19
 
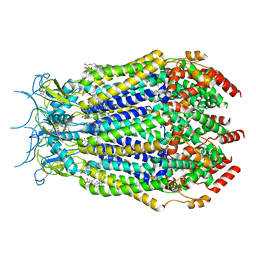 | | SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-2) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 7-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}heptanoic acid, Volume-regulated anion channel subunit LRRC8A | | Authors: | Kern, D.M, Gerber, E.E, Brohawn, S.G. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Small molecule SWELL1 complex induction improves glycemic control and nonalcoholic fatty liver disease in murine Type 2 diabetes.
Nat Commun, 13, 2022
|
|
3LMS
 
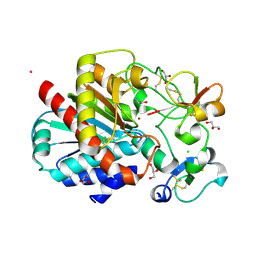 | | Structure of human activated thrombin-activatable fibrinolysis inhibitor, TAFIa, in complex with tick-derived funnelin inhibitor, TCI. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Carboxypeptidase B2, ... | | Authors: | Sanglas, L, Arolas, J.L, Valnickova, Z, Aviles, F.X, Enghild, J.J, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-01 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the molecular inactivation mechanism of human activated thrombin-activatable fibrinolysis inhibitor
J.Thromb.Haemost., 8, 2010
|
|
4RLG
 
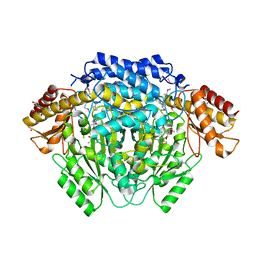 | |
4Q2E
 
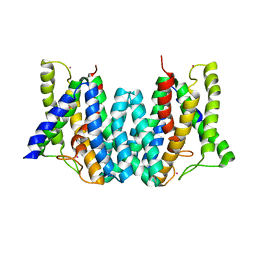 | | CRYSTAL STRUCTURE OF AN INTRAMEMBRANE CDP-DAG SYNTHETASE CENTRAL FOR PHOSPHOLIPID BIOSYNTHESIS (S200C/S258C, active mutant) | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, POTASSIUM ION, ... | | Authors: | Liu, X, Yin, Y, Wu, J, Liu, Z. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis
Nat Commun, 5, 2014
|
|
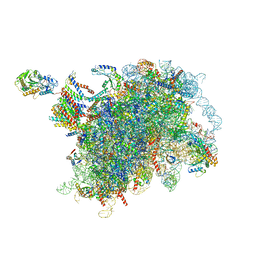
6W6L
 
 | |
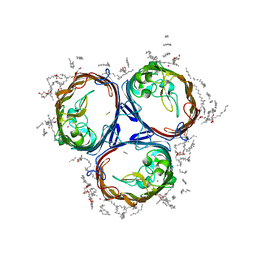
3POX
 
 | |
6F29
 
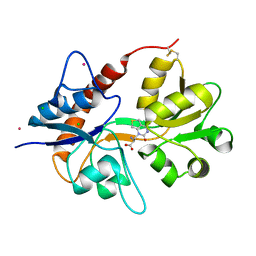 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2-Amino-2-carboxyethyl]-5,7-dihydrothieno[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.6A | | Descriptor: | (2~{S})-2-azanyl-3-[2,4-bis(oxidanylidene)-5,7-dihydrothieno[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2017-11-23 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
7B1S
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum at 0.994-A resolution | | Descriptor: | (2S)-2-{[(2S)-2-{[(2S)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-25 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.992 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
7B2C
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum gassed with xenon | | Descriptor: | (2R)-2-[(2S)-2-[(2S)-2-oxidanylpropoxy]propoxy]propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
5T61
 
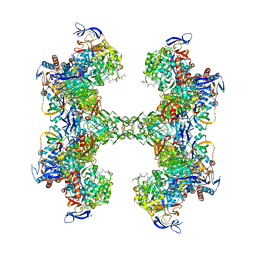 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, TRICLINIC FORM AT 2.55 A | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, HYDROSULFURIC ACID, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
5T5I
 
 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, ORTHORHOMBIC FORM AT 1.9 A | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-19 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
5T5M
 
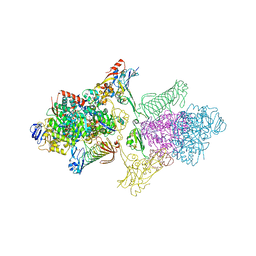 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, TRIGONAL FORM AT 2.5 A. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, GLYCEROL, HYDROSULFURIC ACID, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
3CDM
 
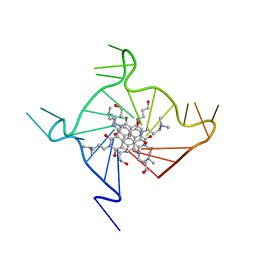 | | Structural adaptation and conservation in quadruplex-drug recognition | | Descriptor: | 2,7-bis[3-(dimethylamino)propyl]-4,9-bis[(3-hydroxypropyl)amino]benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, DNA (5'-D(*DT*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DG)-3'), POTASSIUM ION | | Authors: | Parkinson, G.N, Neidle, S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes
J.Mol.Biol., 381, 2008
|
|
4V7W
 
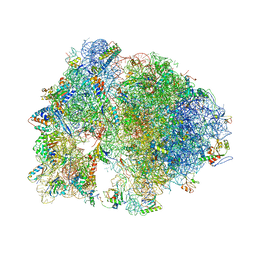 | | Structure of the Thermus thermophilus ribosome complexed with chloramphenicol. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1MAG
 
 | |
3LKI
 
 | |
4A0M
 
 | | CRYSTAL STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE FROM SPINACH IN COMPLEX WITH NAD | | Descriptor: | BETAINE ALDEHYDE DEHYDROGENASE, CHLOROPLASTIC, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Diaz-Sanchez, A.G, Munoz-Clares, R.A. | | Deposit date: | 2011-09-09 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amino Acid Residues Critical for the Specificity for Betaine Aldehyde of the Plant Aldh10 Isoenzyme Involved in the Synthesis of Glycine Betaine.
Plant Physiol., 158, 2012
|
|
5JZX
 
 | | Crystal Structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Mycobacterium tuberculosis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, POTASSIUM ION, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Dharavath, S, Eniyan, K, Bajpai, U, Gourinath, S. | | Deposit date: | 2016-05-17 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine-enolpyruvate reductase (MurB) from Mycobacterium tuberculosis
Biochim. Biophys. Acta, 1866, 2017
|
|
2WME
 
 | | Crystallographic structure of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | BETA-MERCAPTOETHANOL, BETAINE ALDEHYDE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Munoz-Clares, R.A, Horjales, E. | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Ternary Complex of Betaine Aldehyde Dehydrogenase from Pseudomonas Aeruginosa Provides New Insight Into the Reaction Mechanism and Shows a Novel Binding Mode of the 2'- Phosphate of Nadp(+) and a Novel Cation Binding Site.
J.Mol.Biol., 385, 2009
|
|
2XDR
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE MUTANT E252A FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, BETAINE ALDEHYDE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Diaz-Sanchez, A.G, Gonzalez-Segura, L, Rudino-Pinera, E, Lira-Rocha, A, Munoz-Clares, R.A. | | Deposit date: | 2010-05-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | A Novel Cysteine-Nadph Covalent Adduct in Pseudomonas Aeruginosa Betaine Aldehyde Dehydrogenase Suggests Important Roles for the Reduced Nucleotide in the Reaction Mechanism
To be Published
|
|
6ENG
 
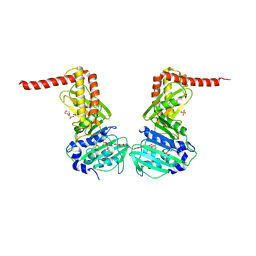 | | Crystal structure of the 43K ATPase domain of Escherichia coli gyrase B in complex with an aminocoumarin | | Descriptor: | CHLORIDE ION, Coumermycin A1, DNA gyrase subunit B, ... | | Authors: | Vanden Broeck, A, McEwen, A.G, Lamour, V. | | Deposit date: | 2017-10-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Gyrase Interaction with Coumermycin A1.
J.Med.Chem., 62, 2019
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
