2KBI
 
 | |
2KUN
 
 | | Three dimensional structure of HuPrP(90-231 M129 Q212P) | | Descriptor: | Major prion protein | | Authors: | Ilc, G, Giachin, G, Jaremko, M, Jaremko, L, Zhukov, I, Plavec, J, Legname, G, Benetti, F. | | Deposit date: | 2010-02-23 | | Release date: | 2010-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human prion protein with the pathological Q212P mutation reveals unique structural features.
Plos One, 5, 2010
|
|
2KS9
 
 | |
2K5O
 
 | |
2LSK
 
 | | C-terminal domain of human REV1 in complex with DNA-polymerase H (eta) | | Descriptor: | DNA polymerase eta, DNA repair protein REV1 | | Authors: | Pozhidaeva, A, Pustovalova, Y, Bezsonova, I, Korzhnev, D. | | Deposit date: | 2012-05-01 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase eta.
Biochemistry, 51, 2012
|
|
2L53
 
 | |
2K8J
 
 | | Solution structure of HCV p7 tm2 | | Descriptor: | p7tm2 | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2008-09-12 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and ion channel activity of the p7 protein from hepatitis C virus.
J.Biol.Chem., 285, 2010
|
|
2HGK
 
 | | Solution NMR Structure of protein YqcC from E. coli: Northeast Structural Genomics Consortium target ER225 | | Descriptor: | Hypothetical protein yqcC | | Authors: | Cort, J.R, Wang, D, Shetty, K, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-27 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein YqcC from E.coli
To be Published
|
|
2JS4
 
 | | Solution NMR Structure of Bordetella bronchiseptica protein BB2007. Northeast Structural Genomics Consortium target BoR54 | | Descriptor: | UPF0434 protein BB2007 | | Authors: | Eletsky, A, Sukumaran, D, Wu, Y, Singarapu, K, Parish, D, Xu, D, Wang, D, Nwosu, C, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Bordetella bronchiseptica protein BB2007.
To be Published
|
|
2JRX
 
 | | Solution NMR structure of protein YejL from E. coli. Northeast Structural Genomics target ER309 | | Descriptor: | UPF0352 protein yejL | | Authors: | Cort, J.R, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Chemical Shift Assignments of E. coli YejL protein.
To be Published
|
|
2JMP
 
 | |
2BAO
 
 | |
2LEZ
 
 | | Solution NMR structure of N-terminal domain of Salmonella effector protein PipB2. Northeast structural genomics consortium (NESG) target stt318a | | Descriptor: | Secreted effector protein pipB2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Daniels, C, Savchenko, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of N-terminal domain of Salmonella effector protein PipB2
To be Published
|
|
2LC3
 
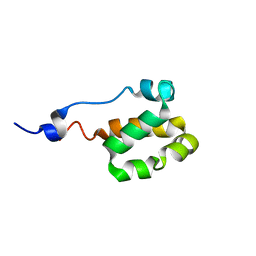 | | Solution NMR structure of a helical bundle domain from human E3 ligase HECTD1. Northeast structural genomics consortium (NESG) target HT6305A | | Descriptor: | E3 ubiquitin-protein ligase HECTD1 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Arrowsmith, C, Dhe-Paganon, S, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-15 | | Release date: | 2011-06-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a helical bundle from the E3 ligase HECTD1
To be Published
|
|
2L8W
 
 | |
2JOC
 
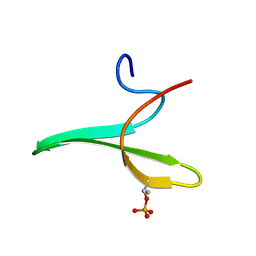 | | Mouse Itch 3rd domain phosphorylated in T30 | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Macias, M.J, Shaw, A.Z, Martin-Malpartida, P, Morales, B, Ruiz, L, Ramirez-Espain, X, Yraola, F, Royo, M. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Structure, 15, 2007
|
|
2JO9
 
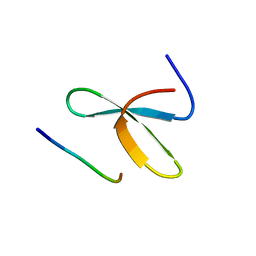 | | Mouse Itch 3rd WW domain complex with the Epstein-Barr virus latent membrane protein 2A derived peptide EEPPPPYED | | Descriptor: | Itchy E3 ubiquitin protein ligase, Latent membrane protein 2 | | Authors: | Macias, M.J, Shaw, A.Z, Martin-Malpartida, P, Morales, B, Ruiz, L, Ramirez-Espain, X, Yraola, F, Royo, M. | | Deposit date: | 2007-03-01 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Structure, 15, 2007
|
|
2CYK
 
 | |
2K85
 
 | | p190-A RhoGAP FF1 domain | | Descriptor: | Glucocorticoid receptor DNA-binding factor 1 | | Authors: | Bonet, R, Ruiz, L, Martin-Malpartida, P, Macias, M. | | Deposit date: | 2008-09-02 | | Release date: | 2009-05-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies on human p190-A RhoGAPFF1 revealed that domain phosphorylation by the PDGF-receptor alpha requires its previous unfolding.
J.Mol.Biol., 389, 2009
|
|
2L0T
 
 | | Solution structure of the complex of ubiquitin and the VHS domain of Stam2 | | Descriptor: | Signal transducing adapter molecule 2, Ubiquitin | | Authors: | Lange, A, Hoeller, D, Wienk, H, Marcillat, O, Lancelin, J, Walker, O. | | Deposit date: | 2010-07-15 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR reveals a different mode of binding of the Stam2 VHS domain to ubiquitin and diubiquitin.
Biochemistry, 50, 2011
|
|
2LP8
 
 | | SOLUTION STRUCTURE OF AN APOPTOSIS ACTIVATING PHOTOSWITCHABLE BAK PEPTIDE BOUND to BCL-XL | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 1 | | Authors: | Wysoczanski, P, Mart, R.J, Loveridge, J.E, Williams, C, Whittaker, S.B.-M, Crump, M.P, Allemann, R.K. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-18 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Photoswitchable Apoptosis Activating Bak Peptide Bound to Bcl-x(L).
J.Am.Chem.Soc., 134, 2012
|
|
2BAU
 
 | |
2K0P
 
 | |
2LET
 
 | | AN 1H NMR DETERMINATION OF THE THREE DIMENSIONAL STRUCTURES OF MIRROR IMAGE FORMS OF A LEU-5 VARIANT OF THE TRYPSIN INHIBITOR ECBALLIUM ELATERIUM (EETI-II) | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Nielsen, K.J, Alewood, D, Andrews, J, Kent, S.B.H, Craik, D.J. | | Deposit date: | 1994-01-04 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | An 1H NMR determination of the three-dimensional structures of mirror-image forms of a Leu-5 variant of the trypsin inhibitor from Ecballium elaterium (EETI-II).
Protein Sci., 3, 1994
|
|
2KLR
 
 | | Solid-state NMR structure of the alpha-crystallin domain in alphaB-crystallin oligomers | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Rajagopal, P, Markovic, S, Bardiaux, B, Kuehne, R, Higman, V.A, Klevit, R.E, van Rossum, B, Oschkinat, H. | | Deposit date: | 2009-07-08 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers.
Nat.Struct.Mol.Biol., 17, 2010
|
|
