1CIB
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH GDP, IMP, HADACIDIN, AND NO3 | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Cashel, M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-03-31 | | Release date: | 2000-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
J.Biol.Chem., 274, 1999
|
|
8R3B
 
 | |
8USK
 
 | |
8USE
 
 | |
8USI
 
 | |
8USL
 
 | |
8USF
 
 | |
8USH
 
 | |
8USJ
 
 | |
8USG
 
 | |
1DJ2
 
 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | Deposit date: | 1999-12-01 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
1P46
 
 | | T4 lysozyme core repacking mutant M106I/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1P7S
 
 | | T4 LYSOZYME CORE REPACKING MUTANT V103I/TA | | Descriptor: | LYSOZYME | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-05-05 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1PQK
 
 | | Repacking of the Core of T4 Lysozyme by Automated Design | | Descriptor: | Lysozyme | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1PQJ
 
 | | T4 LYSOZYME CORE REPACKING MUTANT A111V/CORE10/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | REPACKING THE CORE OF T4 LYSOZYME BY AUTOMATED DESIGN
J.Mol.Biol., 332, 2003
|
|
1PQM
 
 | | T4 Lysozyme Core Repacking Mutant V149I/T152V/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1SOO
 
 | | ADENYLOSUCCINATE SYNTHETASE INHIBITED BY HYDANTOCIDIN 5'-MONOPHOSPHATE | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, BETA-MERCAPTOETHANOL, HYDANTOCIDIN-5'-PHOSPHATE, ... | | Authors: | Cowan-Jacob, S.W. | | Deposit date: | 1996-05-07 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mode of action and the structure of a herbicide in complex with its target: binding of activated hydantocidin to the feedback regulation site of adenylosuccinate synthetase.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1SON
 
 | | ADENYLOSUCCINATE SYNTHETASE IN COMPLEX WITH THE NATURAL FEEDBACK INHIBITOR AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLOSUCCINATE SYNTHETASE, BETA-MERCAPTOETHANOL | | Authors: | Cowan-Jacob, S.W. | | Deposit date: | 1996-05-07 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The mode of action and the structure of a herbicide in complex with its target: binding of activated hydantocidin to the feedback regulation site of adenylosuccinate synthetase.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1CH8
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH A STRINGENT EFFECTOR, PPG2':3'P | | Descriptor: | GUANOSINE 5'-DIPHOSPHATE 2':3'-CYCLIC MONOPHOSPHATE, HADACIDIN, INOSINIC ACID, ... | | Authors: | Hou, Z, Cashel, M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-03-31 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
J.Biol.Chem., 274, 1999
|
|
1DY3
 
 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
2N8D
 
 | |
1OZ0
 
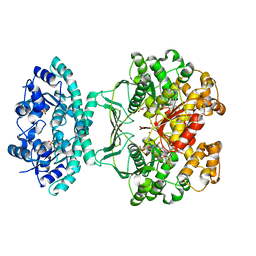 | | CRYSTAL STRUCTURE OF THE HOMODIMERIC BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME AVIAN ATIC IN COMPLEX WITH A MULTISUBSTRATE ADDUCT INHIBITOR BETA-DADF. | | Descriptor: | 2-[4-((2-AMINO-4-OXO-3,4-DIHYDRO-PYRIDO[3,2-D]PYRIMIDIN-6-YLMETHYL)-{3-[5-CARBAMOYL-3-(3,4- DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-3H-IMIDAZOL-4-YL]-ACRYLOYL}-AMINO)-BENZOYLAMINO]- PENTANEDIOIC ACID, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Wolan, D.W, Greasley, S.E, Wall, M.J, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 2003-04-07 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Avian AICAR Transformylase with a Multisubstrate Adduct Inhibitor beta-DADF Identifies the Folate Binding Site.
Biochemistry, 42, 2003
|
|
4C6C
 
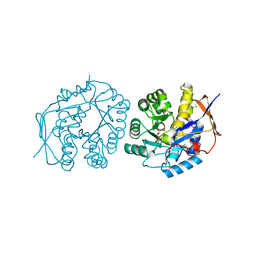 | | Crystal structure of the dihydroorotase domain of human CAD in apo- form obtained recombinantly from HEK293 cells. | | Descriptor: | CAD PROTEIN, FORMIC ACID, ZINC ION | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6O
 
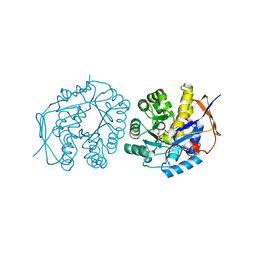 | | Crystal structure of the dihydroorotase domain of human CAD C1613S mutant in apo-form at pH 6.0 | | Descriptor: | CAD PROTEIN, FORMIC ACID, ZINC ION | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6N
 
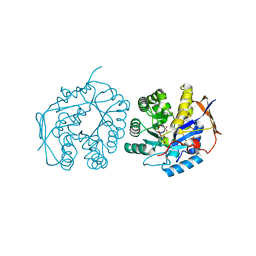 | | Crystal structure of the dihydroorotase domain of human CAD E1637T mutant bound to substrate at pH 6.0 | | Descriptor: | CAD PROTEIN, FORMIC ACID, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
