3SIO
 
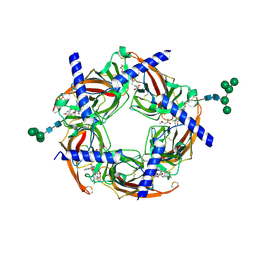 | | Ac-AChBP ligand binding domain (not including beta 9-10 linker) mutated to human alpha-7 nAChR | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemecz, A, Taylor, P.W. | | Deposit date: | 2011-06-19 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Creating an alpha-7 nicotinic acetylcholine recognition domain from the
acetylcholine binding protein: crystallographic and ligand selectivity analyses
J.Biol.Chem., 286, 2011
|
|
1UBS
 
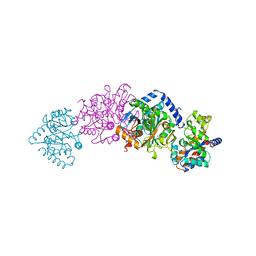 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) WITH A MUTATION OF LYS 87->THR IN THE B SUBUNIT AND IN THE PRESENCE OF LIGAND L-SERINE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, SODIUM ION, ... | | Authors: | Rhee, S, Parris, K, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
2I83
 
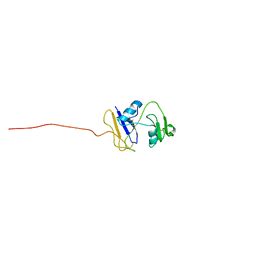 | | hyaluronan-binding domain of CD44 in its ligand-bound form | | Descriptor: | CD44 antigen | | Authors: | Takeda, M, Ogino, S, Umemoto, R, Sakakura, M, Kajiwara, M, Sugahara, K.N, Hayasaka, H, Miyasaka, M, Terasawa, H, Shimada, I. | | Deposit date: | 2006-09-01 | | Release date: | 2006-11-21 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Ligand-induced Structural Changes of the CD44 Hyaluronan-binding Domain Revealed by NMR
J.Biol.Chem., 281, 2006
|
|
5XSV
 
 | |
7WPN
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor and ATP | | Descriptor: | (phenylmethyl) N-[(2R)-2-[2-(4-bromanyl-3-oxidanyl-phenyl)-5-(methylcarbamoyl)benzimidazol-1-yl]-2-(3,4-dimethoxyphenyl)ethyl]carbamate, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPM
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
4UET
 
 | | Diversity in the structures and ligand binding sites among the fatty acid and retinol binding proteins of nematodes revealed by Na-FAR-1 from Necator americanus | | Descriptor: | NEMATODE FATTY ACID RETINOID BINDING PROTEIN | | Authors: | Rey-Burusco, M.F, Ibanez Shimabukuro, M, Griffiths, K, Cooper, A, Kennedy, M.W, Corsico, B, Smith, B.O, Griffiths, K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Diversity in the Structures and Ligand Binding Sites of Nematode Fatty Acid and Retinol Binding Proteins Revealed by Na-Far-1 from Necator Americanus.
Biochem.J., 471, 2015
|
|
4TSE
 
 | |
1KS6
 
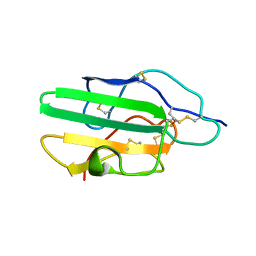 | |
4UM5
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and Phosphate ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UMF
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion, Phosphate ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2V3P
 
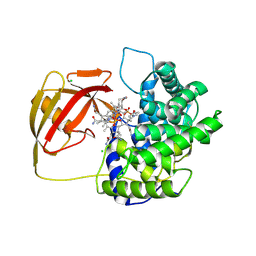 | | Crystallographic analysis of beta-axial ligand substitutions in cobalamin bound to transcobalamin | | Descriptor: | CHLORIDE ION, COBALAMIN, SULFITE ION, ... | | Authors: | Wuerges, J, Geremia, S, Randaccio, L. | | Deposit date: | 2007-06-19 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Vitamin B12 Transport Proteins: Crystallographic Analysis of Beta-Axial Ligand Substitutions in Cobalamin Bound to Transcobalamin.
Iubmb Life, 59, 2007
|
|
4UME
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3S9E
 
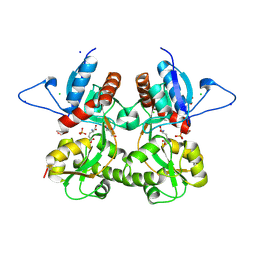 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-06-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
4UMD
 
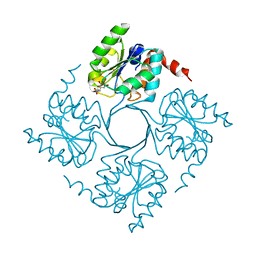 | |
4UM7
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (kdsC) from Moraxella catarrhalis in complex with Magnesium ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8SGE
 
 | | KLHDC2 Kelch Domain with ligand KDRLKZ-1 | | Descriptor: | GLYCEROL, Kelch domain-containing protein 2, [(5P)-5-{3-[(2R)-butan-2-yl]-7-[(2-methoxyethoxy)carbonyl]-2-oxo-5,6,7,8-tetrahydro-1,7-naphthyridin-1(2H)-yl}-2-oxopyridin-1(2H)-yl]acetic acid | | Authors: | Digianantonio, K.M, Bekes, M, Langley, D.R, Zimmerman, K. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Co-opting the E3 ligase KLHDC2 for targeted protein degradation by small molecules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5AVM
 
 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
6LI1
 
 | | Crystal structure of GPR52 ligand free form with flavodoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Flavodoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
1TT1
 
 | |
6L88
 
 | |
4UE1
 
 | | Structure of the stapled peptide YS-01 bound to MDM2 | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE MDM2, YS-01 | | Authors: | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | Deposit date: | 2014-12-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
6LI2
 
 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
9IMO
 
 | | Crystal structure of Tubulin-RB3-TTL-Y12 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-04 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IM5
 
 | | Tubulin-RB3(MUT)-TTL-Y12 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-02 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
