4PYY
 
 | | Crystal structure of human carbonic anhydrase isozyme II with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-03-28 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
5FH6
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with compound 10 | | Descriptor: | (3~{R})-3-(piperidin-1-ylmethyl)-2,3-dihydro-1~{H}-pyrrolo[1,2-a]quinazolin-5-one, Protein polybromo-1 | | Authors: | Tallant, C, Sutherell, C.L, Siejka, P, Krojer, T, Picaud, S, Fonseca, M, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Ley, S.V, Knapp, S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Development of 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-one Inhibitors Targeting Bromodomains within the Switch/Sucrose Nonfermenting Complex.
J.Med.Chem., 59, 2016
|
|
3PE1
 
 | | Crystal structure of human protein kinase CK2 alpha subunit in complex with the inhibitor CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Battistutta, R, Papinutto, E, Lolli, G, Pierre, F, Haddach, M, Ryckman, D.M. | | Deposit date: | 2010-10-25 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer.
Biochemistry, 50, 2011
|
|
3PBG
 
 | | 6-PHOSPHO-BETA-GALACTOSIDASE FORM-C | | Descriptor: | 6-PHOSPHO-BETA-D-GALACTOSIDASE, SULFATE ION | | Authors: | Wiesmann, C, Schulz, G.E. | | Deposit date: | 1997-02-21 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures and mechanism of 6-phospho-beta-galactosidase from Lactococcus lactis.
J.Mol.Biol., 269, 1997
|
|
4F21
 
 | | Crystal structure of carboxylesterase/phospholipase family protein from Francisella tularensis | | Descriptor: | Carboxylesterase/phospholipase family protein, N-((1R,2S)-2-allyl-4-oxocyclobutyl)-4-methylbenzenesulfonamide, bound form | | Authors: | Filippova, E.V, Minasov, G, Kuhn, M, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J.R, Kiryukhina, O, Becker, D.P, Armoush, N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large scale structural rearrangement of a serine hydrolase from Francisella tularensis facilitates catalysis.
J.Biol.Chem., 288, 2013
|
|
5F7R
 
 | |
6D3M
 
 | | FT_T dioxygenase with bound quizalofop | | Descriptor: | (2R)-2-{4-[(6-chloroquinoxalin-2-yl)oxy]phenoxy}propanoic acid, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
3PE2
 
 | | Crystal structure of human protein kinase CK2 in complex with the inhibitor CX-5011 | | Descriptor: | 5-[(3-ethynylphenyl)amino]pyrimido[4,5-c]quinoline-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Battistutta, R, Papinutto, E, Lolli, G, Pierre, F, Haddach, M, Ryckman, D.M. | | Deposit date: | 2010-10-25 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer.
Biochemistry, 50, 2011
|
|
4ZN9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Oxabicyclic Heptene Sulfonate (OBHS) | | Descriptor: | Estrogen receptor, Nuclear receptor-interacting peptide, cyclohexa-2,5-dien-1-yl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands.
Chemmedchem, 7, 2012
|
|
2Q0X
 
 | |
3U8J
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3531 (1-(pyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(pyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
4F2E
 
 | |
5L8B
 
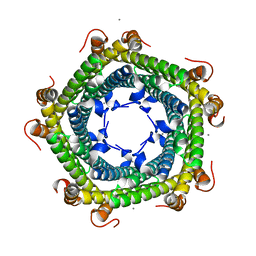 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E62A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
1LU3
 
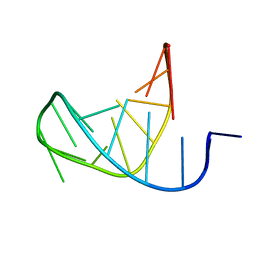 | | Separate Fitting of the Anticodon Loop Region of tRNA (nucleotide 26-42) in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | Descriptor: | PHENYLALANINE TRANSFER RNA | | Authors: | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | Deposit date: | 2002-05-21 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16.799999 Å) | | Cite: | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
6I3A
 
 | | Crystal structure of v22Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | BROMIDE ION, v22Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
4N7I
 
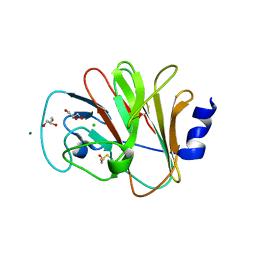 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 | | Descriptor: | BETA-MERCAPTOETHANOL, Butyrophilin subfamily 3 member A1, CHLORIDE ION, ... | | Authors: | Sandstrom, A, Adams, E.J. | | Deposit date: | 2013-10-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4015 Å) | | Cite: | The Intracellular B30.2 Domain of Butyrophilin 3A1 Binds Phosphoantigens to Mediate Activation of Human V gamma 9V delta 2 T Cells.
Immunity, 40, 2014
|
|
3KB3
 
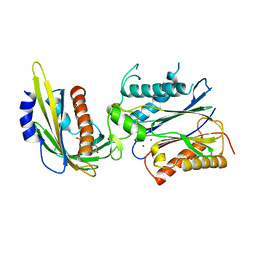 | | Crystal structure of abscisic acid-bound PYL2 in complex with HAB1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Melcher, K, Soon, F.-F, Ng, L.-M, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
6I9I
 
 | |
3PF6
 
 | | The structure of uncharacterized protein PP-LUZ7_gp033 from Pseudomonas phage LUZ7. | | Descriptor: | CHLORIDE ION, hypothetical protein PP-LUZ7_gp033 | | Authors: | Cuff, M.E, Evdokimova, E, Liu, F, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of uncharacterized protein PP-LUZ7_gp033 from Pseudomonas phage LUZ7.
TO BE PUBLISHED
|
|
1C8L
 
 | | SYNERGISTIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG AND CAFFEINE | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, CAFFEINE, GLYCEROL, ... | | Authors: | Tsitsanou, K.E, Skamnaki, V.T, Oikonomakos, N.G. | | Deposit date: | 2000-05-16 | | Release date: | 2000-05-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the synergistic inhibition of glycogen phosphorylase a by caffeine and a potential antidiabetic drug.
Arch.Biochem.Biophys., 384, 2000
|
|
7K6M
 
 | | Crystal structure of PI3Kalpha selective Inhibitor PF-06843195 | | Descriptor: | 2,2-difluoroethyl (3S)-3-{[2'-amino-5-fluoro-2-(morpholin-4-yl)[4,5'-bipyrimidin]-6-yl]amino}-3-(hydroxymethyl)pyrrolidine-1-carboxylate, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
5V3D
 
 | | Crystal structure of fosfomycin resistance protein from Klebsiella pneumoniae with bound fosfomycin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FOSFOMYCIN, ... | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-07 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
5Q05
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methoxyethyl)-4-methylthiophen-2-yl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methoxyethyl)-4-methylthiophen-2-yl]sulfonylurea
To be published
|
|
2OCS
 
 | | The crystal structure of the first PDZ domain of human NHERF-2 (SLC9A3R2) | | Descriptor: | 1,2-ETHANEDIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF2, THIOCYANATE ION | | Authors: | Papagrigoriou, E, Phillips, C, Gileadi, C, Elkins, J, Salah, E, Berridge, G, Savitsky, P, Gorrec, F, Umeano, C, Ugochukwu, E, Gileadi, O, Burgess, N, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the first PDZ domain of human NHERF-2 (SLC9A3R2)
To be Published
|
|
1CA8
 
 | | Thrombin inhibitors with rigid tripeptidyl aldehydes | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3S)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-04-27 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes
Biochemistry, 37, 1998
|
|
