3SGR
 
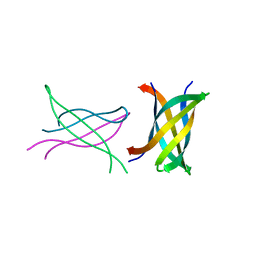 | | Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
3SGX
 
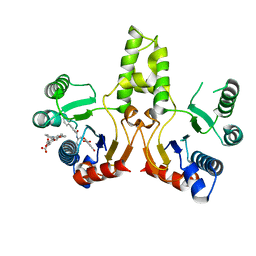 | |
3SHQ
 
 | | Crystal Structure of UBLCP1 | | Descriptor: | MAGNESIUM ION, UBLCP1 | | Authors: | Xiao, J, Engel, J.L. | | Deposit date: | 2011-06-16 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SI0
 
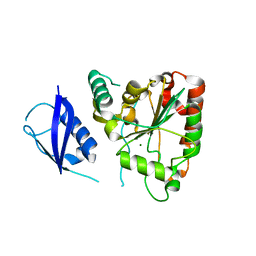
 | | Structure of glycosylated human glutaminyl cyclase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
3SIH
 
 | |
3S6P
 
 | |
3S8P
 
 | | Crystal Structure of the SET Domain of Human Histone-Lysine N-Methyltransferase SUV420H1 In Complex With S-Adenosyl-L-Methionine | | Descriptor: | Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Lam, R, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-30 | | Release date: | 2011-07-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2.
Febs Lett., 587, 2013
|
|
3S97
 
 | | PTPRZ CNTN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Bouyain, S. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2971 Å) | | Cite: | A complex between contactin-1 and the protein tyrosine phosphatase PTPRZ controls the development of oligodendrocyte precursor cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SK6
 
 | |
3SA4
 
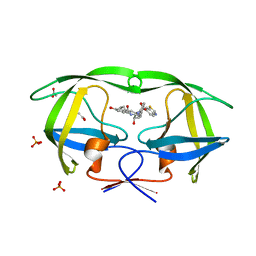 | | Crystal structure of wild-type HIV-1 protease in complex with AF72 | | Descriptor: | ACETATE ION, N-{(2S,3R)-4-[(1,3-benzothiazol-6-ylsulfonyl)(cyclohexylmethyl)amino]-3-hydroxy-1-phenylbutan-2-yl}-3-hydroxybenzamide, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SAC
 
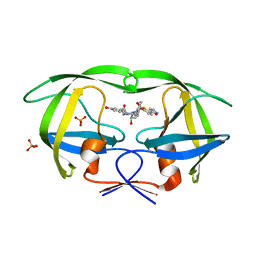 | | Crystal structure of wild-type HIV-1 protease in complex with AF80 | | Descriptor: | 3-hydroxy-N-{(2S,3R)-3-hydroxy-4-[(2-methylpropyl){[5-(1,2-oxazol-5-yl)thiophen-2-yl]sulfonyl}amino]-1-phenylbutan-2-yl}benzamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SL3
 
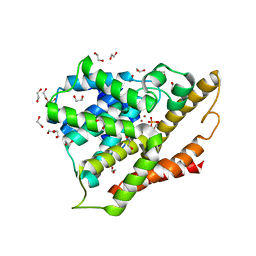 | | Crystal structure of the apo form of the catalytic domain of PDE4D2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SBI
 
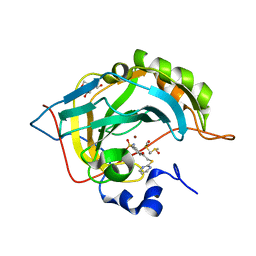 | | Crystal structure of human carbonic anhydrase isozyme II with 4-[(2-pyrimidinylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-06-05 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
3SBU
 
 | |
3SNV
 
 | |
3SO5
 
 | |
3S29
 
 | |
3SH5
 
 | |
3SHZ
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-chloro-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3S4C
 
 | | Lactose phosphorylase in complex with sulfate | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Lactose Phosphorylase, SULFATE ION | | Authors: | Van Hoorebeke, A, Stout, J, Soetaert, W, Van Beeumen, J, Desmet, T, Savvides, S. | | Deposit date: | 2011-05-19 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellobiose phosphorylase: reconstructing the structural itinerary along the catalytic pathway
To be Published
|
|
3SJ7
 
 | |
3S5T
 
 | |
3S5Z
 
 | | Pharmacological Chaperoning in Human alpha-Galactosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, GLYCEROL, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2011-05-23 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The molecular basis of pharmacological chaperoning in human alpha-galactosidase
Chem.Biol., 18, 2011
|
|
3SML
 
 | | Crystal structure of human 14-3-3 sigma C38N/N166H in complex with TASK-3 peptide and stabilizer Fusicoccin A aglycone | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Fusicoccin A aglycone, ... | | Authors: | Anders, C, Schumacher, B, Ottmann, C. | | Deposit date: | 2011-06-28 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface.
Chem. Biol., 20, 2013
|
|
3S6L
 
 | |
