7ZRS
 
 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C2) - composite map | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZUW
 
 | | Structure of RQT (C1) bound to the stalled ribosome in a disome unit from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZPQ
 
 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C1) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
2UZA
 
 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Chabriere, E, Cavazza, C, Contreras-Martel, C, Fontecilla-Camps, J.C. | | Deposit date: | 2007-04-27 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate.
Structure, 14, 2006
|
|
7PP4
 
 | |
7Q59
 
 | |
6Z1L
 
 | |
7ZUX
 
 | | Collided ribosome in a disome unit from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
2FTE
 
 | | Bacteriophage HK97 Expansion Intermediate IV | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
1FMZ
 
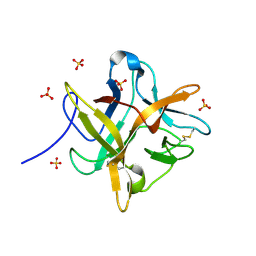 | | CRYSTAL STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14K. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
1FN0
 
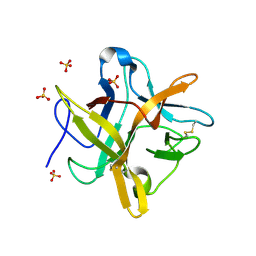 | | STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14D. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
1EYL
 
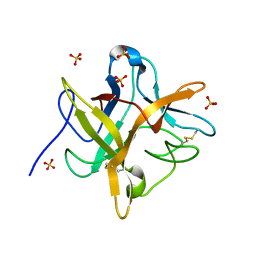 | | STRUCTURE OF A RECOMBINANT WINGED BEAN CHYMOTRYPSIN INHIBITOR | | Descriptor: | CHYMOTRYPSIN INHIBITOR, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Ghosh, S. | | Deposit date: | 2000-05-07 | | Release date: | 2000-05-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
Protein Eng., 14, 2001
|
|
1FXW
 
 | |
2D11
 
 | |
2C3Y
 
 | | CRYSTAL STRUCTURE OF THE RADICAL FORM OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-13 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C3U
 
 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus, Oxygen inhibited form | | Descriptor: | 2-(3-{[4-(HYDROXYAMINO)-2-METHYLPYRIMIDIN-5-YL]METHYL}-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL)ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, IRON/SULFUR CLUSTER, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-12 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C3O
 
 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C42
 
 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-14 | | Release date: | 2006-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C3M
 
 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus | | Descriptor: | CALCIUM ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C3P
 
 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | 1-(2-{(2S,4R,5R)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-[(1S)-1-CARBOXY-1-HYDROXYETHYL]-4-METHYL-1,3-THIAZOLIDIN-5-YL}ETHOXY)-1,1,3,3-TETRAHYDROXY-1LAMBDA~5~-DIPHOSPHOX-1-EN-2-IUM 3-OXIDE, CALCIUM ION, IRON/SULFUR CLUSTER, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2UXN
 
 | | Structural Basis of Histone Demethylation by LSD1 Revealed by Suicide Inactivation | | Descriptor: | CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Szewczuk, L.M, Gocke, C.B, Brautigam, C.A, Tomchick, D.R, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8B0F
 
 | | CryoEM structure of C5b8-CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bubeck, D, Couves, E.C, Gardner, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
7YIG
 
 | | Human KCNH5 pre-open state 2 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily H member 5 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-07-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism underlying delayed rectifying in human voltage-mediated activation Eag2 channel.
Nat Commun, 14, 2023
|
|
7YIE
 
 | | Human KCNH5-closed state 2 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily H member 5 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-07-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism underlying delayed rectifying in human voltage-mediated activation Eag2 channel.
Nat Commun, 14, 2023
|
|
7YIH
 
 | | Human KCNH5 open state | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily H member 5 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-07-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism underlying delayed rectifying in human voltage-mediated activation Eag2 channel.
Nat Commun, 14, 2023
|
|
