3VHR
 
 | | Crystal Structure of capsular polysaccharide assembling protein CapF from Staphylococcus aureus in space group C2221 | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, FORMIC ACID, ZINC ION | | Authors: | Miyafusa, T, Yoshikazu, T, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains.
Biochem.J., 443, 2012
|
|
3VI7
 
 | |
3VK8
 
 | | Crystal structure of DNA-glycosylase bound to DNA containing Thymine glycol | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(CTG)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3'), GLYCEROL, ... | | Authors: | Imamura, K, Averill, A, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-11-10 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA
J.Biol.Chem., 287, 2012
|
|
3VOL
 
 | | X-ray Crystal Structure of PAS-HAMP Aer2 in the CN-bound Form | | Descriptor: | Aerotaxis transducer Aer2, CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Shiro, Y, Aono, S. | | Deposit date: | 2012-01-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for oxygen sensing and signal transduction of the heme-based sensor protein Aer2 from Pseudomonas aeruginosa
Chem.Commun.(Camb.), 48, 2012
|
|
3VOW
 
 | | Crystal Structure of the Human APOBEC3C having HIV-1 Vif-binding Interface | | Descriptor: | CHLORIDE ION, Probable DNA dC->dU-editing enzyme APOBEC-3C, ZINC ION | | Authors: | Kitamura, S, Suzuki, A, Watanabe, N, Iwatani, Y. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The APOBEC3C crystal structure and the interface for HIV-1 Vif binding.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VPL
 
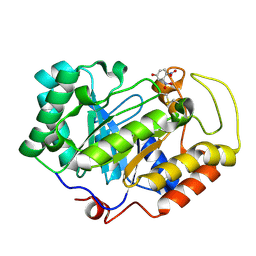 | | Crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 Beta-1,3-xylanase | | Descriptor: | 3,4-dinitrophenol, Beta-1,3-xylanase XYL4, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-3)-2-deoxy-2-fluoro-beta-D-xylopyranose, ... | | Authors: | Watanabe, N, Sakaguchi, K. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 beta-1,3-xylanase at 1.2 A resolution
To be Published
|
|
3VMJ
 
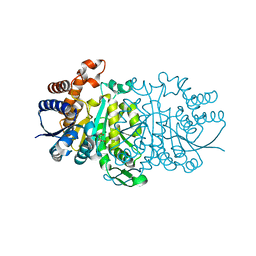 | | 3-isopropylmalate dehydrogenase from Shewanella oneidensis MR-1 | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, CHLORIDE ION, ... | | Authors: | Nagae, T, Watanabe, N. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural analysis of 3-isopropylmalate dehydrogenase from the obligate piezophile Shewanella benthica DB21MT-2 and the nonpiezophile Shewanella oneidensis MR-1
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3VEG
 
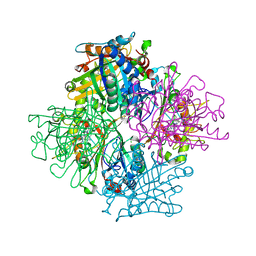 | | Rhodococcus jostii RHA1 DypB R244L variant in complex with heme | | Descriptor: | CHLORIDE ION, DypB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grigg, J.C, Singh, R, Armstrong, Z, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2012-01-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity.
J.Biol.Chem., 287, 2012
|
|
3VEX
 
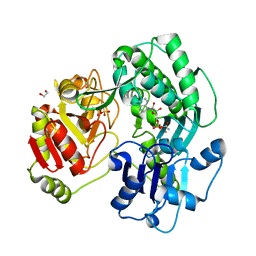 | | Crystal structure of the O-carbamoyltransferase TobZ H14N variant in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, FE (II) ION, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3V88
 
 | | Thaumatin by Classical Hanging Drop Vapour Diffusion after 18.1 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Scudieri, D, Tripathi, S, Pechkova, E, Nicolini, C. | | Deposit date: | 2011-12-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-Blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
CRIT.REV.EUKARYOT.GENE EXPR., 22, 2012
|
|
3VFE
 
 | | Virtual Screening and X-Ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group | | Descriptor: | 4-{[(3R)-3-{[(7-methoxynaphthalen-2-yl)sulfonyl](thiophen-3-ylmethyl)amino}-2-oxopyrrolidin-1-yl]methyl}thiophene-2-carboximidamide, Kallikrein-6 | | Authors: | Chen, X, Zhang, Y, Xia, T, Wang, R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Virtual Screening and X-ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group.
Acs Med.Chem.Lett., 3, 2012
|
|
3VFV
 
 | | crystal structure of HLA B*3508 LPEP-P9Ala, peptide mutant P9-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P9A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3V9L
 
 | |
3VGE
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VL3
 
 | |
3VED
 
 | | Rhodococcus jostii RHA1 DypB D153H variant in complex with heme | | Descriptor: | CHLORIDE ION, DypB, GLYCEROL, ... | | Authors: | Grigg, J.C, Singh, R, Armstrong, Z, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2012-01-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity.
J.Biol.Chem., 287, 2012
|
|
3VLX
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - ligand free form from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3VF9
 
 | |
3VFW
 
 | | crystal structure of HLA B*3508 LPEP-P10Ala, peptide mutant P10-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P10A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VGB
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (GTHase) from Sulfolobus solfataricus KM1 | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VHH
 
 | | Crystal structure of DiMe-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1,3-dimethyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VIB
 
 | |
3VK7
 
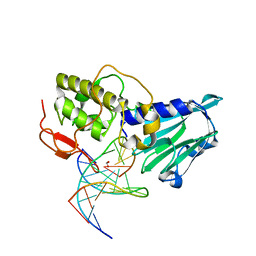 | | Crystal structure of DNA-glycosylase bound to DNA containing 5-Hydroxyuracil | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*GP*AP*CP*GP*TP*GP*GP*AP*CP*G)-3'), FORMIC ACID, ... | | Authors: | Imamura, K, Averill, A, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-11-10 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA
J.Biol.Chem., 287, 2012
|
|
3VKI
 
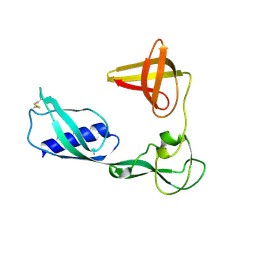 | |
3VL2
 
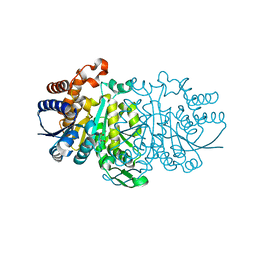 | |
